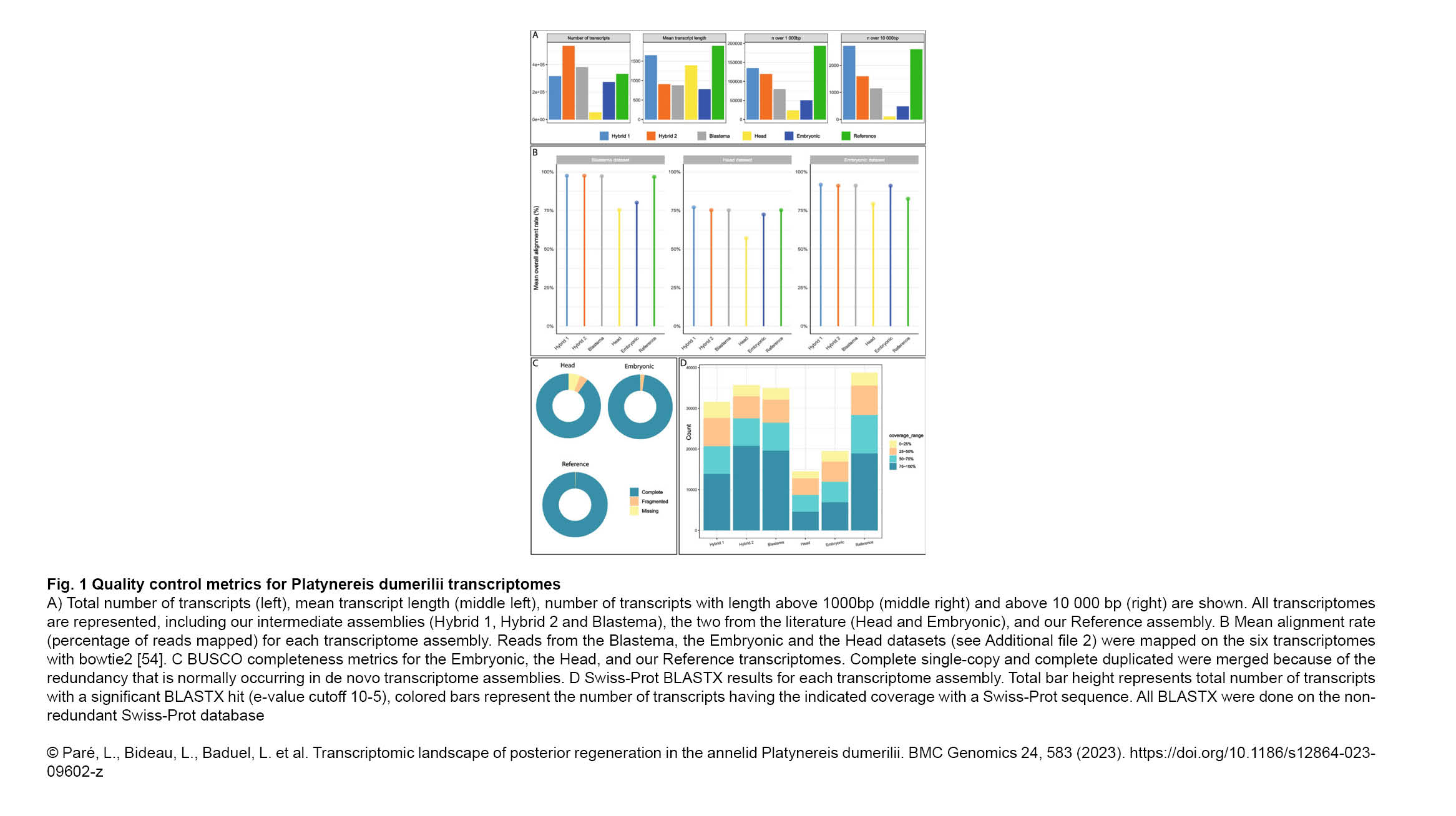
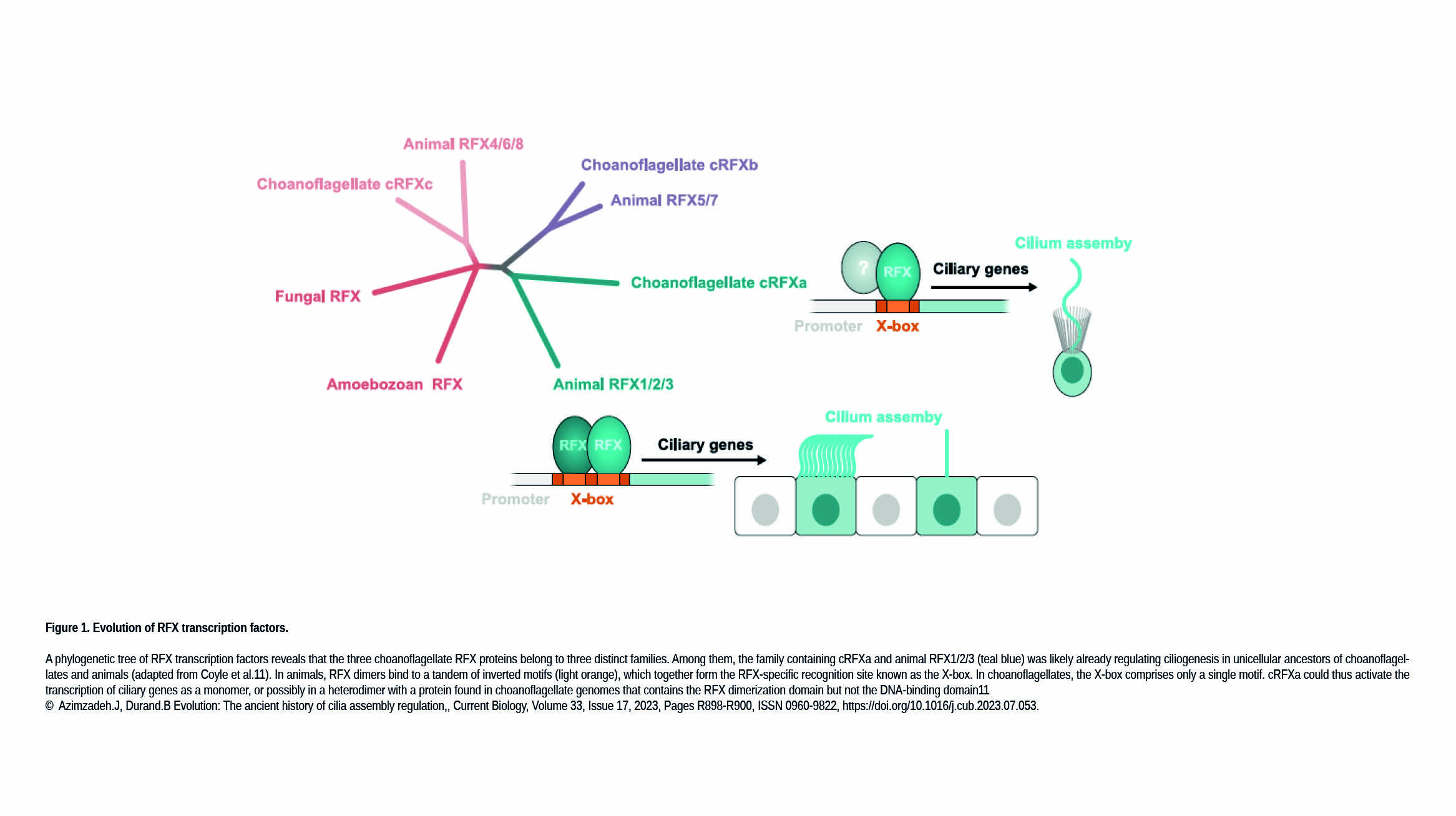
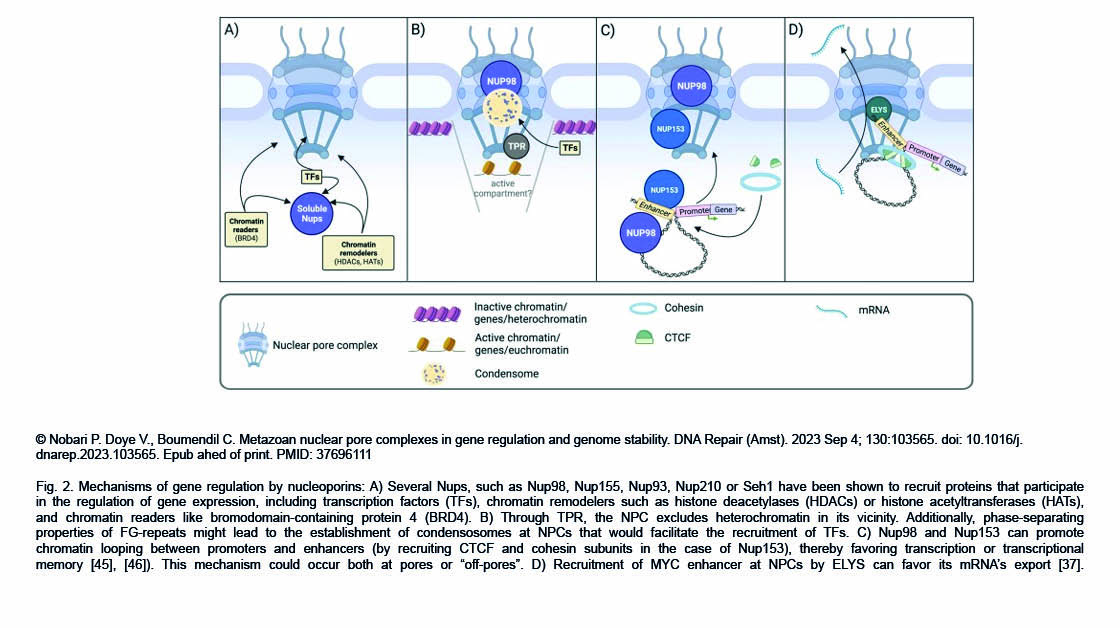
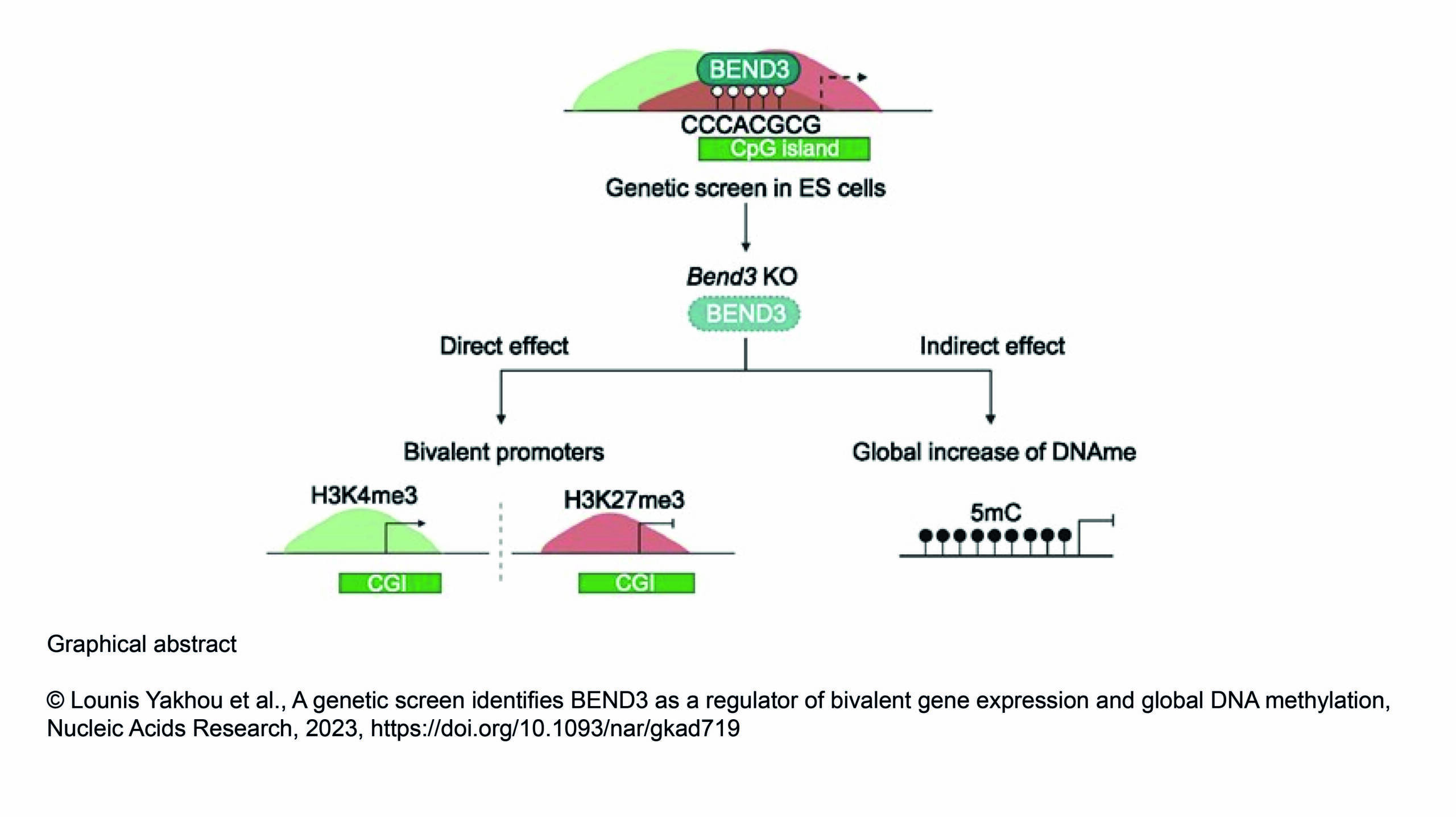
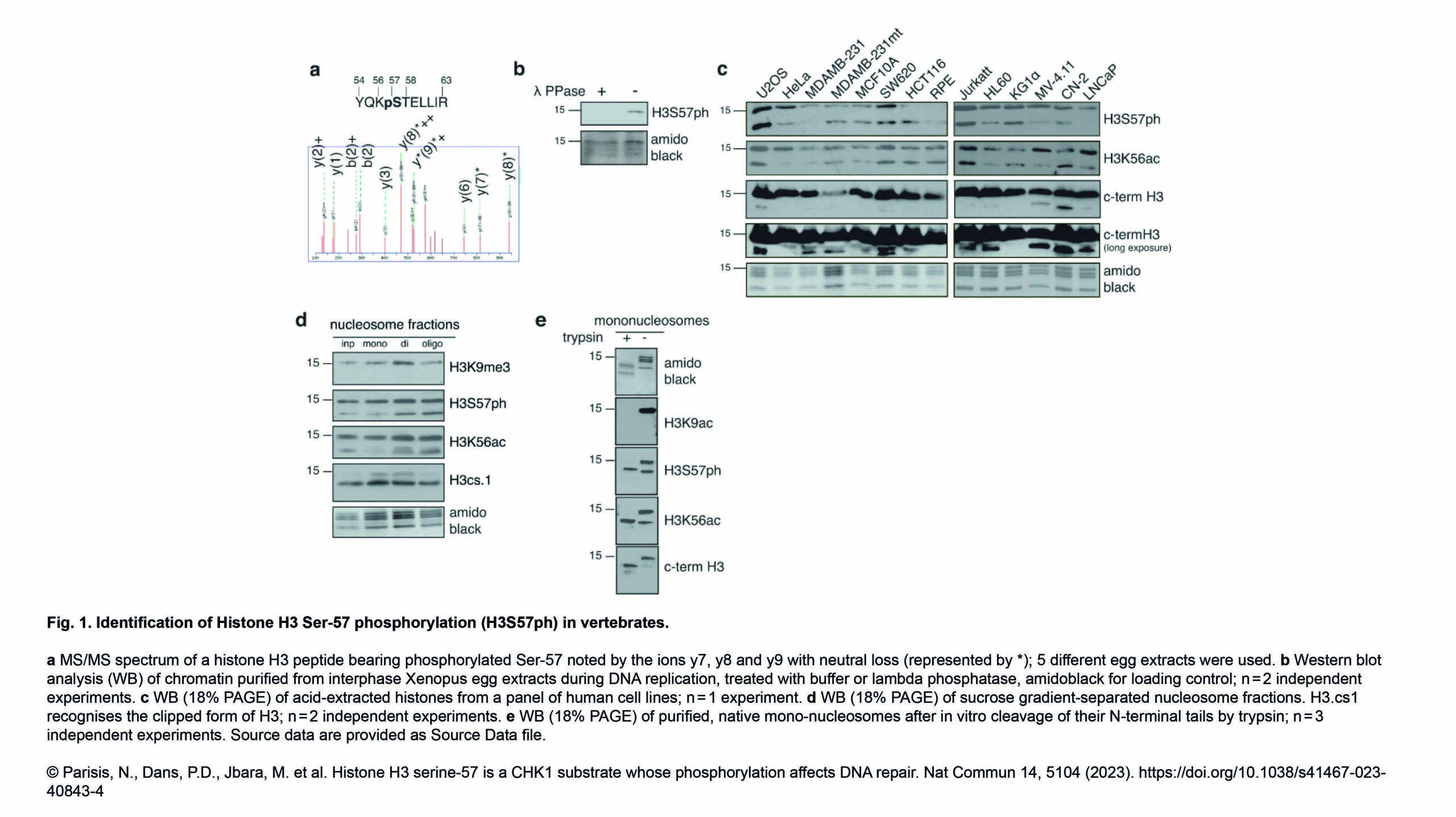
The Gazave Lab recently published a new article in BMC Genomics:
Transcriptomic landscape of posterior regeneration in the annelid Platynereis dumerilii
Abstract:
Background
Restorative regeneration, the capacity to reform a lost body part following amputation or injury, is an important and still poorly understood process in animals. Annelids, or segmented worms, show amazing regenerative capabilities, and as…