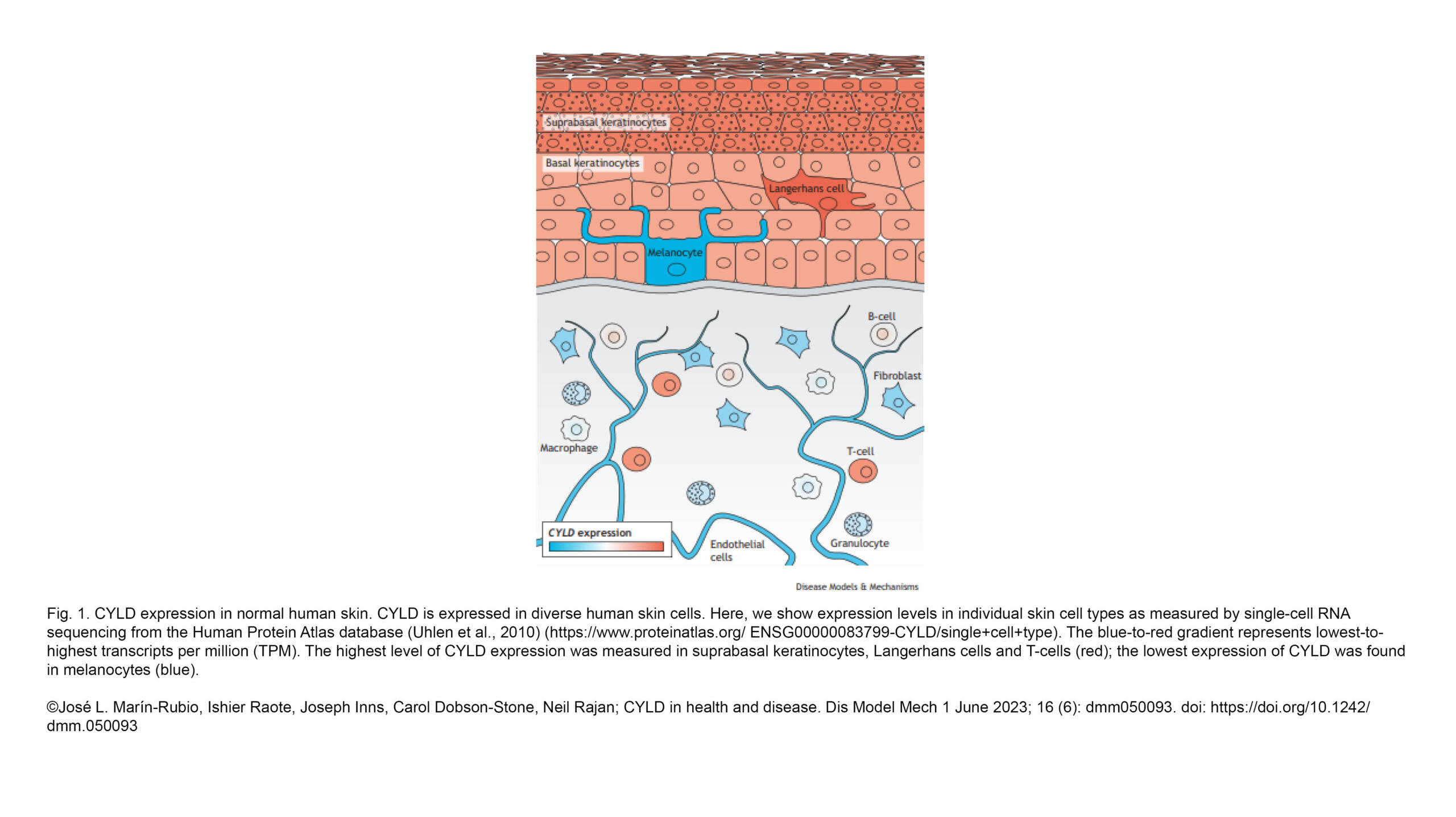
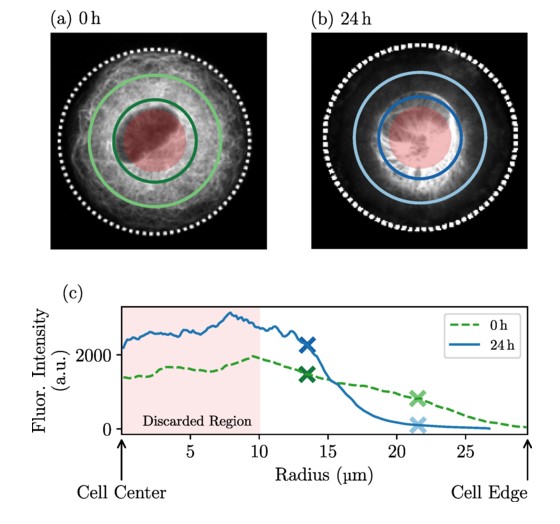
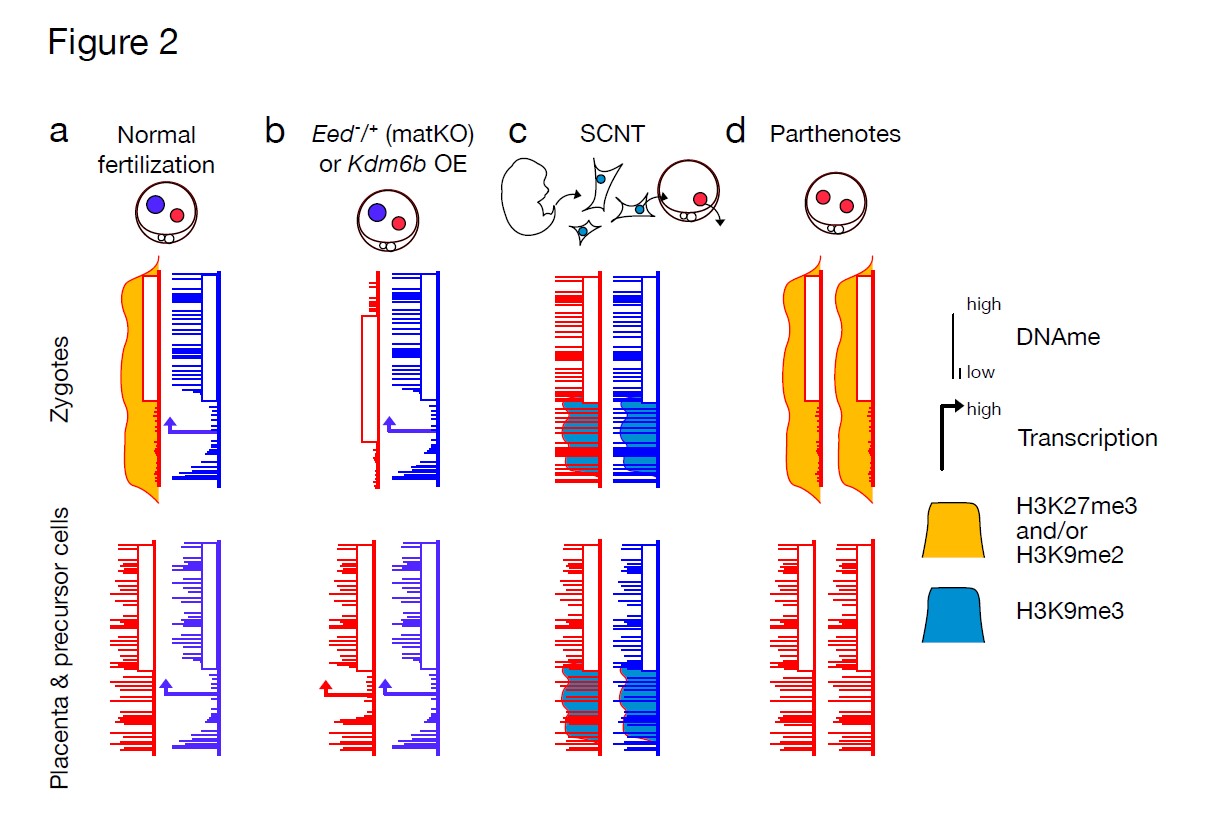
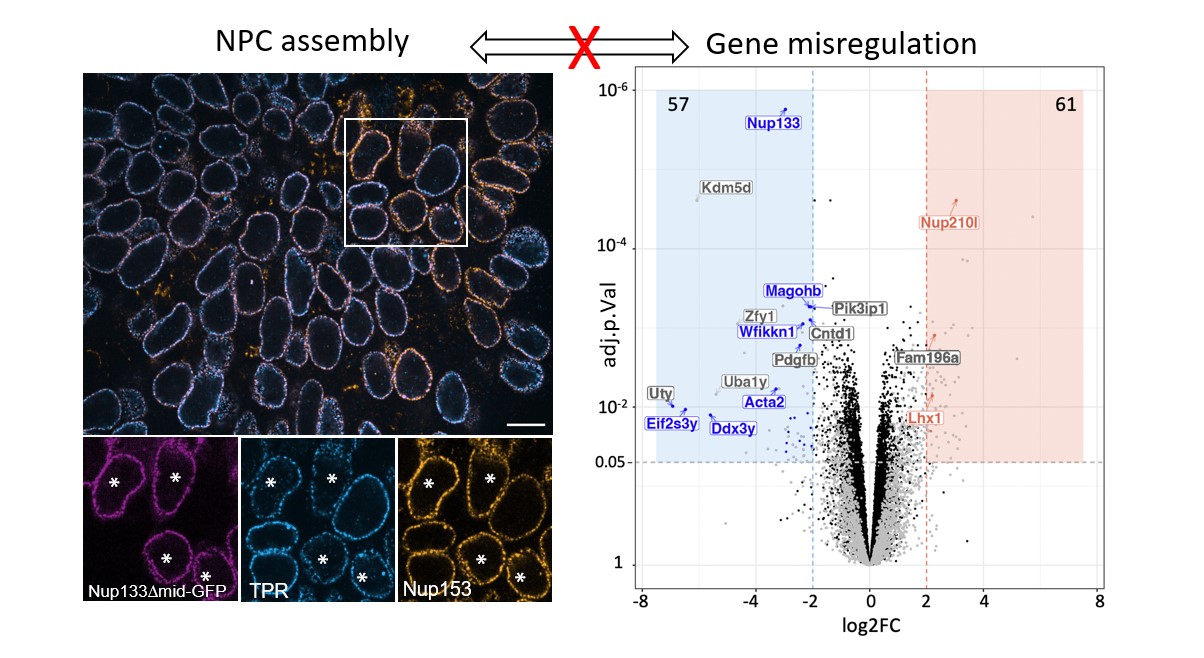
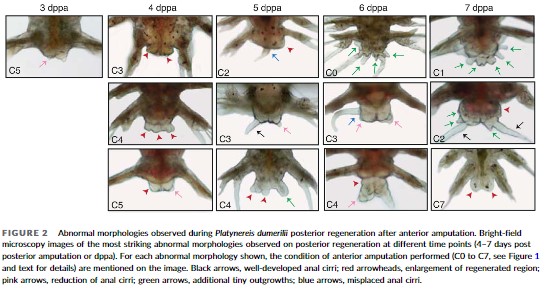
The Raote Lab contributed to the new review published in Disease Models & Mechanisms:
CYLD in health and disease
Abstract:
CYLD lysine 63 deubiquitinase (CYLD) is a ubiquitin hydrolase with important roles in immunity and cancer. Complete CYLD ablation, truncation and expression of alternate isoforms, including short CYLD, drive distinct phenotypes and offer insights into CYLD function in inflammation,…