Nous développons et coordonnons dans l’équipe 3 axes de recherche et un axe de collaborations :
Projet 1 : Dynamique du réplisome : Caractérisation des sites de pauses des ADN polymérases au cours de la réplication dans le génome humain, rôle de l’Hélicase Pif1 et impact sur la stabilité du génome
Nos travaux visent à identifier les mécanismes moléculaires des arrêts de fourches de réplication, en utilisant des cellules humaines. Ces travaux devraient nous permettre de mieux comprendre comment ces défauts de réplication que sont ces arrêts de fourches, peuvent contribuer au développement de cancers associés à une instabilité génomique. La question est maintenant de savoir si les pauses de la réplication sont dues : i) soit à des événements stochastiques, c’est à dire qu’elles se produisent à l’endroit X au moment Y et peuvent varier d’un cycle cellulaire à un autre, ou ii) soit elles se déroulent à une localisation spécifique qui, si elles sont coordonnées avec d’autres processus moléculaires, permettrait d’organiser le génome et de diminuer ainsi le risque d’instabilité génétique.
Pour réaliser cette cartographie des sites de pause, nous posons le postulat que les loci qui entravent la progression des fourches de réplication, présentent une association plus longue avec le réplisome comparée avec les sites où il n’y a pas de pauses. Ces sites de pause doivent donc présenter une forte association avec les ADN polymérases réplicatives. Nous pouvons donc observer la distribution de l’ADN polymérase delta sur le génome dans des cellules asynchrones en cours de réplication avec l’aide d’approches NGS (approche ChIP-seq avec l’anticorps Pol Delta).
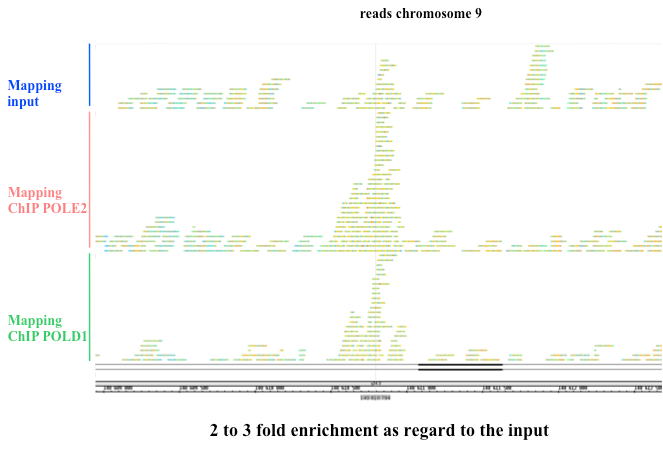 profil obtenu à partir d’un séquençage avec les anticorps anti pol epsilon et delta ainsi que celui obtenu sur l’échantillon Input
profil obtenu à partir d’un séquençage avec les anticorps anti pol epsilon et delta ainsi que celui obtenu sur l’échantillon Input
Nous cherchons aussi à déterminer par des approches NGS et biochimiques le rôle de l’hélicase Pif1 durant la réplication et l’impact d’une dérégulation de cette enzyme sur la stabilité du génome. Durant la phase-S, les fourches de réplication peuvent rencontrer plusieurs types de structures sur la chromatine qui peuvent entraver la progression des fourches. Ces obstacles peuvent avoir plusieurs origines, par exemple : a) la formation d’une structure secondaire de l’ADN, b) la formation d’un complexe ADN-Protéine(s) c) une modification de la chromatine et/ou enfin d) la transcription en elle-même. Les hélicases de la famille Pif1 sont présentes dans pratiquement tous les eucaryotes. Pif1 est nécessaire pour la stabilité de l’ADN mitochondrial. Elle semble aussi intervenir dans la résolution de structures particulières telles les G-quadruplex (expérience in vitro). Enfin Rrm3 (homologue de Pif1) est connu chez S. cerevisae pour faciliter la progression de la fourche à travers les obstacles. Les conflits avec la transcription peuvent avoir deux origines : i) l’association de protéines de la transcription avec la chromatine, ou ii) la formation d’hybrides ARN-ADN issus de la transcription appelés R-loop. Ce projet de recherche consiste donc à caractériser le rôle de Pif1 dans le processus de la réplication et de son impact dans la stabilité du génome. En d’autre terme, nous aimerions savoir si l’absence ou la surexpression de Pif1, induit un stress réplicatif. Nous avons obtenu dans la lignée K562 humaine les clones KO de Pif1 par des approches CRISP/Cas9 mais aussi une lignée de surexpression de Pif1 par des approches PiggyBAC dans laquelle une étiquette HA a été ajoutée à la protéine. Nous avons initié sur ces deux lignées une première expérience de comparaison du timing de réplication
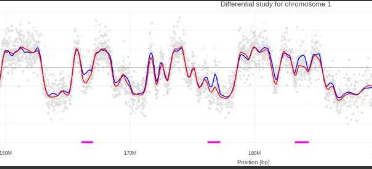
Exemples de profil de timing de réplication obtenus pour la lignée KO Pif1 et la lignée Surexpression de Pif1. En bleu, la lignée WT, en rouge la lignée modifiée génétiquement. Les traits verts indiquent une avancée de timing pour le mutant dans les régions concernées, les traits roses, des retards.
Nous pouvons déjà observer que l’absence ou la surexpression de Pif1 dérégule le timing de réplication. Ces effets sont exacerbés en créant un stress réplicatif supplémentaire avec la présence d’Aphidicoline. Enfin, nous observons un ralentissement de la croissance cellulaire pour ces différentes lignées.
Projet 2 : Impact des inhibiteurs de CDK4-CDK6 sur la réplication et conséquence sur la stabilité du génome
Le palbociclib est une molécule thérapeutique prometteuse dans la lutte contre le cancer, car elle fait partie de la famille des « inhibiteurs de CDK4 et 6 ». Toutefois, des phénomènes de résistance à ces molécules peuvent apparaître chez les patients atteints de cancer. Les cancers dits « négatifs pour la protéine pRb » sont considérés comme résistants à ces molécules : les cellules continuent à proliférer malgré le traitement. Cependant, nous avons montré récemment que ces cellules continuaient à proliférer mais avaient une plus grande difficulté à réaliser la réplication de l’ADN. Cette molécule induit donc un « stress réplicatif ». Aujourd’hui, nous essayons de comprendre à quel niveau cette molécule interfère dans le processus de la réplication. Par des approches de peignage moléculaire, nous avons observé une diminution du nombre d’origines de réplication actives, mais aussi une accélération de la vitesse de synthèse du réplisome. Ce traitement a un effet sur la composition moléculaire du Pré-complexe d’Initiation des origines de réplication.
Projet 3 : impact de la Parylation sur le contrôle de la réplication
La PARylation, via l’enzyme PARP1 et le NAD+, est le processus moléculaire permettant la synthèse d’une chaîne à plusieurs branches d’ADP-Ribose (PAR) au niveau d’une protéine (c’est une modification post-traductionnelle) ou au niveau de l’ADN. Cette chaîne poly-ADP-ribosique permet de recruter d’autres protéines qui possèdent des motifs peptidiques de liaison au PAR. Ainsi la PARylation peut être impliquée dans plusieurs processus moléculaires tels que : le remodelage de la chromatine, la transcription, la réparation de l’ADN et le cycle cellulaire. L’implication de PARP1 dans la réparation de l’ADN est la mieux décrite aujourd’hui. Brièvement, PARP1 agit comme un détecteur de cassure car elle peut interagir avec l’ADN grâce aux deux motifs en doigt de zinc que possède cette protéine dans un de ses domaines. Avec cette interaction, PARP1 va ensuite synthétiser une chaîne PAR permettant par la suite le recrutement sur le site de cassure des protéines impliquées dans la réparation de l’ADN.
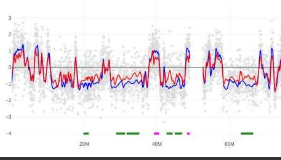
L’implication de PARP1 dans la réplication de l’ADN est très partiellement établie mais les mécanismes moléculaires associés ne sont pas encore clairs. Il a été observé, dans des anciens travaux par des approches de microscopie à fluorescence (Coll et al., 1997), que PARP1 semble être co-localisé aux lieux de synthèse de l’ADN mais aussi à l’initiation de cette synthèse durant la phase S du cycle cellulaire. D’autres travaux semblent montrer la présence de PARP1 sur des réplisomes arrêtés (Ronson et al., 2018). Nous avons réalisé des expériences où les cellules sont traitées par PARPi ou PARGi, inhibiteur de l’enzyme PARP et PARG respectivement. Nous avons observé que les cellules ont un timing de réplication fortement perturbé. Une cinétique d’incorporation de l’EdU a été réalisée pour voir l’effet de ce type de traitement sur la synthèse de l’ADN et sur les protéines du stress réplicatif. Nous observons à la deuxième heure de traitement, une diminution d’incorporation d’EdU. Concernant les protéines de stress réplicatif, nous observons une fois de plus dès la deuxième heure, l’apparition de la phosphorylation de ChK1, protéine majeure de la réponse aux stress réplicatifs. L’association de ces données préliminaires indique clairement le rôle direct de la PARylation dans le processus de la réplication.
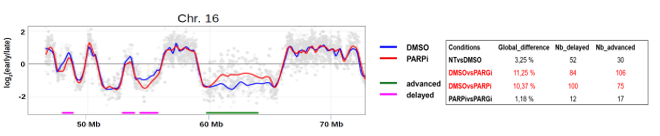
L’inhibition de PARP par l’Olaparib et de PARG par un inhibiteur spécifique a un impact sur le programme temporel de réplication dans les cellules K562. (gauche) Exemple d’un profil de timing de réplication visualisé via START-R. Les points gris correspondent aux sondes qui couvrent chacune une région du chromosome humain 16 (hg18). L’axe des y indique la réplication précoce (valeurs positives) et la réplication tardive (valeurs négatives). (droite)Analyse des différences dans le timing de réplication entre les conditions indiquées. Le nombre de régions à réplication retardée ou avancée est également indiqué.
Projet 4 : Analyse du programme temporel de réplication et collaborations.
Nous avons réalisé une App-Shiny pour étudier le programme temporel de la réplication sur tout le génome (doi : 10.5281/zenodo.3243339 et https://thomasdenecker.github.io/START-R/index.html ).
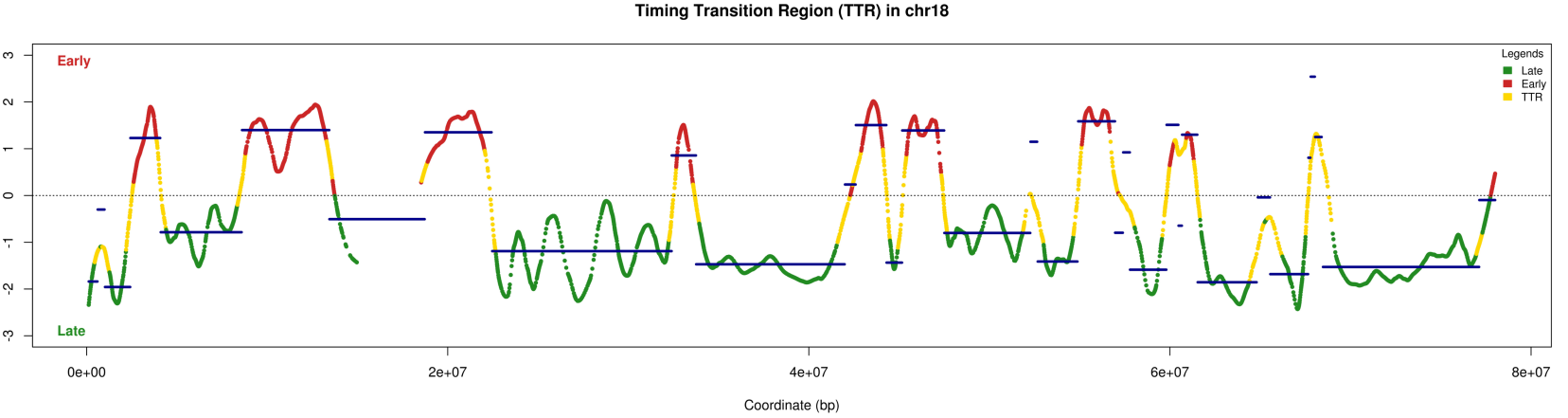
copie d’écran du logiciel
Responsable
Jean-Charles CADORET,
Enseignant-chercheur,
CADORET LAB+33 (0)1 57 27 80 73, bureau 254B
Membres
Giuseppe BALDACCI,
Enseignant-chercheur émérite,
CADORET LAB+33 (0)1 57 27 80 73, bureau RB05B
Samuel BENSOUSSAN,
Stagiaire,
CADORET LAB Lina-Marie BRIU,
Doctorante,
CADORET LAB+33 (0)1 57 27 80 74, bureau 254B
Chrystelle MARIC ANTOINAT,
Chercheur,
CADORET LAB+33 (0)1 57 27 80 73, bureau 254B
Pour contacter un membre de l’équipe par mail : prenom.nom@ijm.fr
- Barbara Ben Yamin, Sana Ahmed-Seghir, Junya Tomida, Emmanuelle Despras, Caroline Pouvelle, Andrey Iurchenko, Jordane Goulas, Raphael Corre, Quentin Delacour, Nathalie Droin, Philippe Dessen, Didier Goidin, Sabine S. Lange, Maria Teresa Mitjavila-Garcia, Giuseppe Baldacci, Sergey Nikolaev, Jean-Charles Cadoret*, Richard D Wood* and Patricia L Kannouche*. DNA polymerase zeta contributes to heterochromatin replication to prevent genome instability. EMBO J. 2021 Nov 2;40(21):e104543. *Co-last authors
- Courtot L, Bournique E, Maric C, Guitton-Sert L, Madrid-Mencía M, Pancaldi V, Jean-Charles Cadoret*, Hoffmann JS*, Bergoglio V*. Low Replicative Stress Triggers Cell-Type Specific Inheritable Advanced Replication timing. Int J Mol Sci. 2021 May 7;22(9):4959. doi: 10.3390/ijms22094959. PMID: 34066960; PMCID: PMC8125030. * co-senior and co-corresponding authors
- Djihad Hadjadj, Thomas Denecker, Eva Guérin, Su-Jung Kim, Fabien Fauchereau, Giuseppe Baldacci, Chrystelle Maric, Jean-Charles Cadoret. Efficient, quick and easy-to-use DNA replication timing analysis with START-R suite. NAR genomics & bioinformatics, 2020 journal 2, vol2.
- Djihad Hadjadj, Su-jung Kim, Thomas Denecker, Laura Ben Driss, Jean-Charles Cadoret, Chrystelle Maric, Giuseppe Baldacci, Fabien Fauchereau. A hypothesis-driven approach identifies CDK4 and CDK6 inhibitors as candidate drugs for treatments of adrenocortical carcinomas. Aging, Albany NY). 2017 Dec 26;9(12):2695-2716
- Julien Brustel, Nina Kirstein, Fanny Izard, Stanimir Dulev, Charlotte Grimaud, Chrystelle Cayrou, Gunnar Schotta, Marcel Mechali, Giuseppe Baldacci, Claude Sardet, Nizar N. Batada, Jean-Charles Cadoret*, Aloys Schepers* & Eric Julien*. Histone H4K20 tri-methylation at late-firing origins ensures timely heterochromatin replication. EMBO J. 2017 Sep 15;36(18):2726-2741, * co-senior and co-corresponding authors & Commentaire dans F1000
- Fernandez-Vidal, Anne *; Guitton-Sert*, Laure; Jean-Charles Cadoret*; et al. A role for DNA polymerase theta in the timing of DNA replication. Nature Communications Volume: 5 Article Number:4285 JUL 2014 * first co-authors, Selected for a research highlight in Nature Reviews Molecular Cell Biology 15, 499 5)
- Briu LM, Maric C, Jean-Charles Cadoret. Replication Stress, Genomic Instability, and Replication timing: A Complex Relationship. Int J Mol Sci. 2021 Apr 30;22(9):4764. doi: 10.3390/ijms22094764. PMID: 33946274; PMCID: PMC8125245.
logiciels : logiciel START-R : https://github.com/thomasdenecker/START-R
Publications
2913254
WM7T2BK4
1
apa
50
date
desc
8554
https://www.ijm.fr/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3Afalse%2C%22meta%22%3A%7B%22request_last%22%3A0%2C%22request_next%22%3A0%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%22PBYQZXTL%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22G%5Cu00f6der%20et%20al.%22%2C%22parsedDate%22%3A%222024-06-12%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EG%26%23xF6%3Bder%2C%20A.%2C%20Maric%2C%20C.%20A.%2C%20Rainey%2C%20M.%20D.%2C%20O%26%23×2019%3BConnor%2C%20A.%2C%20Cazzaniga%2C%20C.%2C%20Shamavu%2C%20D.%2C%20Cadoret%2C%20J.-C.%2C%20%26amp%3B%20Santocanale%2C%20C.%20%282024%29.%20DBF4%2C%20not%20DRF1%2C%20is%20the%20crucial%20regulator%20of%20CDC7%20kinase%20at%20replication%20forks.%20%3Ci%3EJournal%20of%20Cell%20Biology%3C%5C%2Fi%3E%2C%20%3Ci%3E223%3C%5C%2Fi%3E%288%29%2C%20e202402144.%20%3Ca%20class%3D%27zp-ItemURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202402144%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202402144%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22DBF4%2C%20not%20DRF1%2C%20is%20the%20crucial%20regulator%20of%20CDC7%20kinase%20at%20replication%20forks%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anja%22%2C%22lastName%22%3A%22G%5Cu00f6der%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Chrystelle%20Antoinat%22%2C%22lastName%22%3A%22Maric%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michael%20D.%22%2C%22lastName%22%3A%22Rainey%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Aisling%22%2C%22lastName%22%3A%22O%5Cu2019Connor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Chiara%22%2C%22lastName%22%3A%22Cazzaniga%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Daniel%22%2C%22lastName%22%3A%22Shamavu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Charles%22%2C%22lastName%22%3A%22Cadoret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Corrado%22%2C%22lastName%22%3A%22Santocanale%22%7D%5D%2C%22abstractNote%22%3A%22CDC7%20kinase%20is%20crucial%20for%20DNA%20replication%20initiation%20and%20is%20involved%20in%20fork%20processing%20and%20replication%20stress%20response.%20Human%20CDC7%20requires%20the%20binding%20of%20either%20DBF4%20or%20DRF1%20for%20its%20activity.%20However%2C%20it%20is%20unclear%20whether%20the%20two%20regulatory%20subunits%20target%20CDC7%20to%20a%20specific%20set%20of%20substrates%2C%20thus%20having%20different%20biological%20functions%2C%20or%20if%20they%20act%20redundantly.%20Using%20genome%20editing%20technology%2C%20we%20generated%20isogenic%20cell%20lines%20deficient%20in%20either%20DBF4%20or%20DRF1%3A%20these%20cells%20are%20viable%20but%20present%20signs%20of%20genomic%20instability%2C%20indicating%20that%20both%20can%20independently%20support%20CDC7%20for%20bulk%20DNA%20replication.%20Nonetheless%2C%20DBF4-deficient%20cells%20show%20altered%20replication%20efficiency%2C%20partial%20deficiency%20in%20MCM%20helicase%20phosphorylation%2C%20and%20alterations%20in%20the%20replication%20timing%20of%20discrete%20genomic%20regions.%20Notably%2C%20we%20find%20that%20CDC7%20function%20at%20replication%20forks%20is%20entirely%20dependent%20on%20DBF4%20and%20not%20on%20DRF1.%20Thus%2C%20DBF4%20is%20the%20primary%20regulator%20of%20CDC7%20activity%2C%20mediating%20most%20of%20its%20functions%20in%20unperturbed%20DNA%20replication%20and%20upon%20replication%20interference.%22%2C%22date%22%3A%222024-06-12%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%2210.1083%5C%2Fjcb.202402144%22%2C%22ISSN%22%3A%220021-9525%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202402144%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222024-06-20T12%3A03%3A15Z%22%7D%7D%2C%7B%22key%22%3A%22VHX2IWRE%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Lebdy%20et%20al.%22%2C%22parsedDate%22%3A%222023-11-15%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ELebdy%2C%20R.%2C%20Canut%2C%20M.%2C%20Patouillard%2C%20J.%2C%20Cadoret%2C%20J.-C.%2C%20Letessier%2C%20A.%2C%20Ammar%2C%20J.%2C%20Basbous%2C%20J.%2C%20Urbach%2C%20S.%2C%20Miotto%2C%20B.%2C%20Constantinou%2C%20A.%2C%20Abou%20Merhi%2C%20R.%2C%20%26amp%3B%20Ribeyre%2C%20C.%20%282023%29.%20The%20nucleolar%20protein%20GNL3%20prevents%20resection%20of%20stalled%20replication%20forks.%20%3Ci%3EEMBO%20Reports%3C%5C%2Fi%3E%2C%20%3Ci%3En%5C%2Fa%3C%5C%2Fi%3E%28n%5C%2Fa%29%2C%20e57585.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.15252%5C%2Fembr.202357585%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.15252%5C%2Fembr.202357585%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20nucleolar%20protein%20GNL3%20prevents%20resection%20of%20stalled%20replication%20forks%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rana%22%2C%22lastName%22%3A%22Lebdy%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marine%22%2C%22lastName%22%3A%22Canut%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julie%22%2C%22lastName%22%3A%22Patouillard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Charles%22%2C%22lastName%22%3A%22Cadoret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne%22%2C%22lastName%22%3A%22Letessier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Josiane%22%2C%22lastName%22%3A%22Ammar%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jihane%22%2C%22lastName%22%3A%22Basbous%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Serge%22%2C%22lastName%22%3A%22Urbach%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Benoit%22%2C%22lastName%22%3A%22Miotto%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Angelos%22%2C%22lastName%22%3A%22Constantinou%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Raghida%22%2C%22lastName%22%3A%22Abou%20Merhi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cyril%22%2C%22lastName%22%3A%22Ribeyre%22%7D%5D%2C%22abstractNote%22%3A%22Abstract%20Faithful%20DNA%20replication%20requires%20specific%20proteins%20that%20protect%20replication%20forks%20and%20so%20prevent%20the%20formation%20of%20DNA%20lesions%20that%20may%20damage%20the%20genome.%20Identification%20of%20new%20proteins%20involved%20in%20this%20process%20is%20essential%20to%20understand%20how%20DNA%20lesions%20accumulate%20in%20cancer%20cells%20and%20how%20they%20tolerate%20them.%20Here%2C%20we%20show%20that%20human%20GNL3%5C%2Fnucleostemin%2C%20a%20GTP-binding%20protein%20localized%20mostly%20in%20the%20nucleolus%20and%20highly%20expressed%20in%20cancer%20cells%2C%20prevents%20nuclease-dependent%20resection%20of%20nascent%20DNA%20in%20response%20to%20replication%20stress.%20We%20demonstrate%20that%20inhibiting%20origin%20firing%20reduces%20resection.%20This%20suggests%20that%20the%20heightened%20replication%20origin%20activation%20observed%20upon%20GNL3%20depletion%20largely%20drives%20the%20observed%20DNA%20resection%20probably%20due%20to%20the%20exhaustion%20of%20the%20available%20RPA%20pool.%20We%20show%20that%20GNL3%20and%20DNA%20replication%20initiation%20factor%20ORC2%20interact%20in%20the%20nucleolus%20and%20that%20the%20concentration%20of%20GNL3%20in%20the%20nucleolus%20is%20required%20to%20limit%20DNA%20resection.%20We%20propose%20that%20the%20control%20of%20origin%20firing%20by%20GNL3%20through%20the%20sequestration%20of%20ORC2%20in%20the%20nucleolus%20is%20critical%20to%20prevent%20nascent%20DNA%20resection%20in%20response%20to%20replication%20stress.%22%2C%22date%22%3A%222023-11-15%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%2210.15252%5C%2Fembr.202357585%22%2C%22ISSN%22%3A%221469-221X%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.embopress.org%5C%2Fdoi%5C%2Ffull%5C%2F10.15252%5C%2Fembr.202357585%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222023-11-23T09%3A56%3A42Z%22%7D%7D%2C%7B%22key%22%3A%22TMYQ7WLG%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kim%20et%20al.%22%2C%22parsedDate%22%3A%222023-01%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKim%2C%20S.-J.%2C%20Maric%2C%20C.%2C%20Briu%2C%20L.-M.%2C%20Fauchereau%2C%20F.%2C%20Baldacci%2C%20G.%2C%20Debatisse%2C%20M.%2C%20Koundrioukoff%2C%20S.%2C%20%26amp%3B%20Cadoret%2C%20J.-C.%20%282023%29.%20Firing%20of%20Replication%20Origins%20Is%20Disturbed%20by%20a%20CDK4%5C%2F6%20Inhibitor%20in%20a%20pRb-Independent%20Manner.%20%3Ci%3EInternational%20Journal%20of%20Molecular%20Sciences%3C%5C%2Fi%3E%2C%20%3Ci%3E24%3C%5C%2Fi%3E%2813%29%2C%2010629.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fijms241310629%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fijms241310629%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Firing%20of%20Replication%20Origins%20Is%20Disturbed%20by%20a%20CDK4%5C%2F6%20Inhibitor%20in%20a%20pRb-Independent%20Manner%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Su-Jung%22%2C%22lastName%22%3A%22Kim%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Chrystelle%22%2C%22lastName%22%3A%22Maric%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lina-Marie%22%2C%22lastName%22%3A%22Briu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fabien%22%2C%22lastName%22%3A%22Fauchereau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Giuseppe%22%2C%22lastName%22%3A%22Baldacci%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michelle%22%2C%22lastName%22%3A%22Debatisse%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phane%22%2C%22lastName%22%3A%22Koundrioukoff%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Charles%22%2C%22lastName%22%3A%22Cadoret%22%7D%5D%2C%22abstractNote%22%3A%22Over%20the%20last%20decade%2C%20CDK4%5C%2F6%20inhibitors%20%28palbociclib%2C%20ribociclib%20and%20abemaciclib%29%20have%20emerged%20as%20promising%20anticancer%20drugs.%20Numerous%20studies%20have%20demonstrated%20that%20CDK4%5C%2F6%20inhibitors%20efficiently%20block%20the%20pRb-E2F%20pathway%20and%20induce%20cell%20cycle%20arrest%20in%20pRb-proficient%20cells.%20Based%20on%20these%20studies%2C%20the%20inhibitors%20have%20been%20approved%20by%20the%20FDA%20for%20treatment%20of%20advanced%20hormonal%20receptor%20%28HR%29%20positive%20breast%20cancers%20in%20combination%20with%20hormonal%20therapy.%20However%2C%20some%20evidence%20has%20recently%20shown%20unexpected%20effects%20of%20the%20inhibitors%2C%20underlining%20a%20need%20to%20characterize%20the%20effects%20of%20CDK4%5C%2F6%20inhibitors%20beyond%20pRb.%20Our%20study%20demonstrates%20how%20palbociclib%20impairs%20origin%20firing%20in%20the%20DNA%20replication%20process%20in%20pRb-deficient%20cell%20lines.%20Strikingly%2C%20despite%20the%20absence%20of%20pRb%2C%20cells%20treated%20with%20palbociclib%20synthesize%20less%20DNA%20while%20showing%20no%20cell%20cycle%20arrest.%20Furthermore%2C%20this%20CDK4%5C%2F6%20inhibitor%20treatment%20disturbs%20the%20temporal%20program%20of%20DNA%20replication%20and%20reduces%20the%20density%20of%20replication%20forks.%20Cells%20treated%20with%20palbociclib%20show%20a%20defect%20in%20the%20loading%20of%20the%20Pre-initiation%20complex%20%28Pre-IC%29%20proteins%20on%20chromatin%2C%20indicating%20a%20reduced%20initiation%20of%20DNA%20replication.%20Our%20findings%20highlight%20hidden%20effects%20of%20palbociclib%20on%20the%20dynamics%20of%20DNA%20replication%20and%20of%20its%20cytotoxic%20consequences%20on%20cell%20viability%20in%20the%20absence%20of%20pRb.%20This%20study%20provides%20a%20potential%20therapeutic%20application%20of%20palbociclib%20in%20combination%20with%20other%20drugs%20to%20target%20genomic%20instability%20in%20pRB-deficient%20cancers.%22%2C%22date%22%3A%222023%5C%2F1%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.3390%5C%2Fijms241310629%22%2C%22ISSN%22%3A%221422-0067%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.mdpi.com%5C%2F1422-0067%5C%2F24%5C%2F13%5C%2F10629%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222023-07-20T12%3A06%3A14Z%22%7D%7D%2C%7B%22key%22%3A%22XBLQIXJK%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Adam%20et%20al.%22%2C%22parsedDate%22%3A%222022-02-01%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EAdam%2C%20Y.%2C%20Brezellec%2C%20P.%2C%20Espinosa%2C%20E.%2C%20Besombes%2C%20A.%2C%20Naquin%2C%20D.%2C%20Paly%2C%20E.%2C%20Possoz%2C%20C.%2C%20van%20Dijk%2C%20E.%2C%20Barre%2C%20F.-X.%2C%20%26amp%3B%20Ferat%2C%20J.-L.%20%282022%29.%20Plesiomonas%20shigelloides%2C%20an%20Atypical%20Enterobacterales%20with%20a%20Vibrio-Related%20Secondary%20Chromosome.%20%3Ci%3EGenome%20Biology%20and%20Evolution%3C%5C%2Fi%3E%2C%20%3Ci%3E14%3C%5C%2Fi%3E%282%29%2C%20evac011.%20%3Ca%20class%3D%27zp-ItemURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fgbe%5C%2Fevac011%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fgbe%5C%2Fevac011%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Plesiomonas%20shigelloides%2C%20an%20Atypical%20Enterobacterales%20with%20a%20Vibrio-Related%20Secondary%20Chromosome%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yazid%22%2C%22lastName%22%3A%22Adam%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pierre%22%2C%22lastName%22%3A%22Brezellec%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elena%22%2C%22lastName%22%3A%22Espinosa%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Amelie%22%2C%22lastName%22%3A%22Besombes%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Delphine%22%2C%22lastName%22%3A%22Naquin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Evelyne%22%2C%22lastName%22%3A%22Paly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Christophe%22%2C%22lastName%22%3A%22Possoz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Erwin%22%2C%22lastName%22%3A%22van%20Dijk%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Francois-Xavier%22%2C%22lastName%22%3A%22Barre%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Luc%22%2C%22lastName%22%3A%22Ferat%22%7D%5D%2C%22abstractNote%22%3A%22About%2010%25%20of%20bacteria%20have%20a%20multichromosome%20genome%20with%20a%20primary%20replicon%20of%20bacterial%20origin%2C%20called%20the%20chromosome%2C%20and%20other%20replicons%20of%20plasmid%20origin%2C%20the%20chromids.%20Studies%20on%20multichromosome%20bacteria%20revealed%20potential%20points%20of%20coordination%20between%20the%20replication%5C%2Fsegregation%20of%20chromids%20and%20the%20progression%20of%20the%20cell%20cycle.%20For%20example%2C%20replication%20of%20the%20chromid%20of%20Vibrionales%20%28called%20Chr2%29%20is%20initiated%20upon%20duplication%20of%20a%20sequence%20carried%20by%20the%20primary%20chromosome%20%28called%20Chr1%29%2C%20in%20such%20a%20way%20that%20replication%20of%20both%20replicons%20is%20completed%20synchronously.%20Also%2C%20Chr2%20uses%20the%20Chr1%20as%20a%20scaffold%20for%20its%20partition%20in%20the%20daughter%20cells.%20How%20many%20of%20the%20features%20detected%20so%20far%20are%20required%20for%20the%20proper%20integration%20of%20a%20secondary%20chromosome%20in%20the%20cell%20cycle%3F%20How%20many%20more%20features%20remain%20to%20be%20discovered%3F%20We%20hypothesized%20that%20critical%20features%20for%20the%20integration%20of%20the%20replication%5C%2Fsegregation%20of%20a%20given%20chromid%20within%20the%20cell%20cycle%20program%20would%20be%20conserved%20independently%20of%20the%20species%20in%20which%20the%20chromid%20has%20settled.%20Hence%2C%20we%20searched%20for%20a%20chromid%20related%20to%20that%20found%20in%20Vibrionales%20outside%20of%20this%20order.%20We%20identified%20one%20in%20Plesiomonas%20shigelloides%2C%20an%20aquatic%20and%20pathogenic%20enterobacterium%20that%20diverged%20early%20within%20the%20clade%20of%20Enterobacterales.%20Our%20results%20suggest%20that%20the%20chromids%20present%20in%20P.%5Cu00a0shigelloides%20and%20Vibrionales%20derive%20from%20a%20common%20ancestor.%20We%20initiated%20in%20silico%20genomic%20and%20proteomic%20comparative%20analyses%20of%20P.%5Cu00a0shigelloides%2C%20Vibrionales%2C%20and%20Enterobacterales%20that%20enabled%20us%20to%20establish%20a%20list%20of%20features%20likely%20involved%20in%20the%20maintenance%20of%20the%20chromid%20within%20the%20host%20cell%20cycle.%22%2C%22date%22%3A%222022-02-01%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%2210.1093%5C%2Fgbe%5C%2Fevac011%22%2C%22ISSN%22%3A%221759-6653%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fgbe%5C%2Fevac011%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222023-05-17T11%3A44%3A07Z%22%7D%7D%2C%7B%22key%22%3A%228NFPCU87%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Ben%20Yamin%20et%20al.%22%2C%22parsedDate%22%3A%222021-11-02%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EBen%20Yamin%2C%20B.%2C%20Ahmed-Seghir%2C%20S.%2C%20Tomida%2C%20J.%2C%20Despras%2C%20E.%2C%20Pouvelle%2C%20C.%2C%20Yurchenko%2C%20A.%2C%20Goulas%2C%20J.%2C%20Corre%2C%20R.%2C%20Delacour%2C%20Q.%2C%20Droin%2C%20N.%2C%20Dessen%2C%20P.%2C%20Goidin%2C%20D.%2C%20Lange%2C%20S.%20S.%2C%20Bhetawal%2C%20S.%2C%20Mitjavila-Garcia%2C%20M.%20T.%2C%20Baldacci%2C%20G.%2C%20Nikolaev%2C%20S.%2C%20Cadoret%2C%20J.%20C.%2C%20Wood%2C%20R.%20D.%2C%20%26amp%3B%20Kannouche%2C%20P.%20L.%20%282021%29.%20DNA%20polymerase%20zeta%20contributes%20to%20heterochromatin%20replication%20to%20prevent%20genome%20instability.%20%3Ci%3EThe%20EMBO%20Journal%3C%5C%2Fi%3E%2C%20%3Ci%3E40%3C%5C%2Fi%3E%2821%29%2C%20e104543.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.15252%5C%2Fembj.2020104543%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.15252%5C%2Fembj.2020104543%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22DNA%20polymerase%20zeta%20contributes%20to%20heterochromatin%20replication%20to%20prevent%20genome%20instability%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Barbara%22%2C%22lastName%22%3A%22Ben%20Yamin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sana%22%2C%22lastName%22%3A%22Ahmed-Seghir%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Junya%22%2C%22lastName%22%3A%22Tomida%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Emmanuelle%22%2C%22lastName%22%3A%22Despras%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Caroline%22%2C%22lastName%22%3A%22Pouvelle%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Andrey%22%2C%22lastName%22%3A%22Yurchenko%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jordane%22%2C%22lastName%22%3A%22Goulas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Raphael%22%2C%22lastName%22%3A%22Corre%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Quentin%22%2C%22lastName%22%3A%22Delacour%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nathalie%22%2C%22lastName%22%3A%22Droin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Philippe%22%2C%22lastName%22%3A%22Dessen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Didier%22%2C%22lastName%22%3A%22Goidin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sabine%20S.%22%2C%22lastName%22%3A%22Lange%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sarita%22%2C%22lastName%22%3A%22Bhetawal%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Maria%20Teresa%22%2C%22lastName%22%3A%22Mitjavila-Garcia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Giuseppe%22%2C%22lastName%22%3A%22Baldacci%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sergey%22%2C%22lastName%22%3A%22Nikolaev%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean%20Charles%22%2C%22lastName%22%3A%22Cadoret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Richard%20D.%22%2C%22lastName%22%3A%22Wood%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Patricia%20L.%22%2C%22lastName%22%3A%22Kannouche%22%7D%5D%2C%22abstractNote%22%3A%22The%20DNA%20polymerase%20zeta%20%28Pol%5Cu03b6%29%20plays%20a%20critical%20role%20in%20bypassing%20DNA%20damage.%20REV3L%2C%20the%20catalytic%20subunit%20of%20Pol%5Cu03b6%2C%20is%20also%20essential%20in%20mouse%20embryonic%20development%20and%20cell%20proliferation%20for%20reasons%20that%20remain%20incompletely%20understood.%20In%20this%20study%2C%20we%20reveal%20that%20REV3L%20protein%20interacts%20with%20heterochromatin%20components%20including%20repressive%20histone%20marks%20and%20localizes%20in%20pericentromeric%20regions%20through%20direct%20interaction%20with%20HP1%20dimer.%20We%20demonstrate%20that%20Pol%5Cu03b6%5C%2FREV3L%20ensures%20progression%20of%20replication%20forks%20through%20difficult-to-replicate%20pericentromeric%20heterochromatin%2C%20thereby%20preventing%20spontaneous%20chromosome%20break%20formation.%20We%20also%20find%20that%20Rev3l-deficient%20cells%20are%20compromised%20in%20the%20repair%20of%20heterochromatin-associated%20double-stranded%20breaks%2C%20eliciting%20deletions%20in%20late-replicating%20regions.%20Lack%20of%20REV3L%20leads%20to%20further%20consequences%20that%20may%20be%20ascribed%20to%20heterochromatin%20replication%20and%20repair-associated%20functions%20of%20Pol%5Cu03b6%2C%20with%20a%20disruption%20of%20the%20temporal%20replication%20program%20at%20specific%20loci.%20This%20is%20correlated%20with%20changes%20in%20epigenetic%20landscape%20and%20transcriptional%20control%20of%20developmentally%20regulated%20genes.%20These%20results%20reveal%20a%20new%20function%20of%20Pol%5Cu03b6%20in%20preventing%20chromosome%20instability%20during%20replication%20of%20heterochromatic%20regions.%22%2C%22date%22%3A%222021-11-02%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.15252%5C%2Fembj.2020104543%22%2C%22ISSN%22%3A%221460-2075%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A09Z%22%7D%7D%2C%7B%22key%22%3A%22AP6VR24W%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kychygina%20et%20al.%22%2C%22parsedDate%22%3A%222021-06-23%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKychygina%2C%20A.%2C%20Dall%26%23×2019%3BOsto%2C%20M.%2C%20Allen%2C%20J.%20A.%20M.%2C%20Cadoret%2C%20J.-C.%2C%20Piras%2C%20V.%2C%20Pickett%2C%20H.%20A.%2C%20%26amp%3B%20Crabbe%2C%20L.%20%282021%29.%20Progerin%20impairs%203D%20genome%20organization%20and%20induces%20fragile%20telomeres%20by%20limiting%20the%20dNTP%20pools.%20%3Ci%3EScientific%20Reports%3C%5C%2Fi%3E%2C%20%3Ci%3E11%3C%5C%2Fi%3E%281%29%2C%2013195.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41598-021-92631-z%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41598-021-92631-z%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Progerin%20impairs%203D%20genome%20organization%20and%20induces%20fragile%20telomeres%20by%20limiting%20the%20dNTP%20pools%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anna%22%2C%22lastName%22%3A%22Kychygina%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marina%22%2C%22lastName%22%3A%22Dall%5Cu2019Osto%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Joshua%20A.%20M.%22%2C%22lastName%22%3A%22Allen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Charles%22%2C%22lastName%22%3A%22Cadoret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vincent%22%2C%22lastName%22%3A%22Piras%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hilda%20A.%22%2C%22lastName%22%3A%22Pickett%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laure%22%2C%22lastName%22%3A%22Crabbe%22%7D%5D%2C%22abstractNote%22%3A%22Chromatin%20organization%20within%20the%20nuclear%20volume%20is%20essential%20to%20regulate%20many%20aspects%20of%20its%20function%20and%20to%20safeguard%20its%20integrity.%20A%20key%20player%20in%20this%20spatial%20scattering%20of%20chromosomes%20is%20the%20nuclear%20envelope%20%28NE%29.%20The%20NE%20tethers%20large%20chromatin%20domains%20through%20interaction%20with%20the%20nuclear%20lamina%20and%20other%20associated%20proteins.%20This%20organization%20is%20perturbed%20in%20cells%20from%20Hutchinson%5Cu2013Gilford%20progeria%20syndrome%20%28HGPS%29%2C%20a%20genetic%20disorder%20characterized%20by%20premature%20aging%20features.%20Here%2C%20we%20show%20that%20HGPS-related%20lamina%20defects%20trigger%20an%20altered%203D%20telomere%20organization%20with%20increased%20contact%20sites%20between%20telomeres%20and%20the%20nuclear%20lamina%2C%20and%20an%20altered%20telomeric%20chromatin%20state.%20The%20genome-wide%20replication%20timing%20signature%20of%20these%20cells%20is%20perturbed%2C%20with%20a%20shift%20to%20earlier%20replication%20for%20regions%20that%20normally%20replicate%20late.%20As%20a%20consequence%2C%20we%20detected%20a%20higher%20density%20of%20replication%20forks%20traveling%20simultaneously%20on%20DNA%20fibers%2C%20which%20relies%20on%20limiting%20cellular%20dNTP%20pools%20to%20support%20processive%20DNA%20synthesis.%20Remarkably%2C%20increasing%20dNTP%20levels%20in%20HGPS%20cells%20rescued%20fragile%20telomeres%2C%20and%20improved%20the%20replicative%20capacity%20of%20the%20cells.%20Our%20work%20highlights%20a%20functional%20connection%20between%20NE%20dysfunction%20and%20telomere%20homeostasis%20in%20the%20context%20of%20premature%20aging.%22%2C%22date%22%3A%222021-06-23%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41598-021-92631-z%22%2C%22ISSN%22%3A%222045-2322%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.nature.com%5C%2Farticles%5C%2Fs41598-021-92631-z%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222023-05-17T11%3A39%3A53Z%22%7D%7D%2C%7B%22key%22%3A%22FNKQXCL4%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Marsin%20et%20al.%22%2C%22parsedDate%22%3A%222021-06-21%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EMarsin%2C%20S.%2C%20Adam%2C%20Y.%2C%20Cargemel%2C%20C.%2C%20Andreani%2C%20J.%2C%20Baconnais%2C%20S.%2C%20Legrand%2C%20P.%2C%20Li%26%23xA0%3Bde%26%23xA0%3Bla%26%23xA0%3BSierra-Gallay%2C%20I.%2C%20Humbert%2C%20A.%2C%20Aumont-Nicaise%2C%20M.%2C%20Velours%2C%20C.%2C%20Ochsenbein%2C%20F.%2C%20Durand%2C%20D.%2C%20Le%26%23xA0%3BCam%2C%20E.%2C%20Walbott%2C%20H.%2C%20Possoz%2C%20C.%2C%20Quevillon-Cheruel%2C%20S.%2C%20%26amp%3B%20Ferat%2C%20J.-L.%20%282021%29.%20Study%20of%20the%20DnaB%3ADciA%20interplay%20reveals%20insights%20into%20the%20primary%20mode%20of%20loading%20of%20the%20bacterial%20replicative%20helicase.%20%3Ci%3ENucleic%20Acids%20Research%3C%5C%2Fi%3E%2C%20%3Ci%3E49%3C%5C%2Fi%3E%2811%29%2C%206569%26%23×2013%3B6586.%20%3Ca%20class%3D%27zp-ItemURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkab463%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkab463%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Study%20of%20the%20DnaB%3ADciA%20interplay%20reveals%20insights%20into%20the%20primary%20mode%20of%20loading%20of%20the%20bacterial%20replicative%20helicase%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phanie%22%2C%22lastName%22%3A%22Marsin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yazid%22%2C%22lastName%22%3A%22Adam%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Claire%22%2C%22lastName%22%3A%22Cargemel%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jessica%22%2C%22lastName%22%3A%22Andreani%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sonia%22%2C%22lastName%22%3A%22Baconnais%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pierre%22%2C%22lastName%22%3A%22Legrand%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ines%22%2C%22lastName%22%3A%22Li%5Cu00a0de%5Cu00a0la%5Cu00a0Sierra-Gallay%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Adeline%22%2C%22lastName%22%3A%22Humbert%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Magali%22%2C%22lastName%22%3A%22Aumont-Nicaise%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Christophe%22%2C%22lastName%22%3A%22Velours%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fran%5Cu00e7oise%22%2C%22lastName%22%3A%22Ochsenbein%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Dominique%22%2C%22lastName%22%3A%22Durand%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eric%22%2C%22lastName%22%3A%22Le%5Cu00a0Cam%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22H%5Cu00e9l%5Cu00e8ne%22%2C%22lastName%22%3A%22Walbott%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Christophe%22%2C%22lastName%22%3A%22Possoz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sophie%22%2C%22lastName%22%3A%22Quevillon-Cheruel%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Luc%22%2C%22lastName%22%3A%22Ferat%22%7D%5D%2C%22abstractNote%22%3A%22Replicative%20helicases%20are%20essential%20proteins%20that%20unwind%20DNA%20in%20front%20of%20replication%20forks.%20Their%20loading%20depends%20on%20accessory%20proteins%20and%20in%20bacteria%2C%20DnaC%20and%20DnaI%20are%20well%20characterized%20loaders.%20However%2C%20most%20bacteria%20do%20not%20express%20either%20of%20these%20two%20proteins.%20Instead%2C%20they%20are%20proposed%20to%20rely%20on%20DciA%2C%20an%20ancestral%20protein%20unrelated%20to%20DnaC%5C%2FI.%20While%20the%20DciA%20structure%20from%20Vibrio%20cholerae%20shares%20no%20homology%20with%20DnaC%2C%20it%20reveals%20similarities%20with%20DnaA%20and%20DnaX%2C%20two%20proteins%20involved%20during%20replication%20initiation.%20As%20other%20bacterial%20replicative%20helicases%2C%20VcDnaB%20adopts%20a%20toroid-shaped%20homo-hexameric%20structure%2C%20but%20with%20a%20slightly%20open%20dynamic%20conformation%20in%20the%20free%20state.%20We%20show%20that%20VcDnaB%20can%20load%20itself%20on%20DNA%20in%20vitro%20and%20that%20VcDciA%20stimulates%20this%20function%2C%20resulting%20in%20an%20increased%20DNA%20unwinding.%20VcDciA%20interacts%20with%20VcDnaB%20with%20a%203%5C%2F6%20stoichiometry%20and%20we%20show%20that%20a%20determinant%20residue%2C%20which%20discriminates%20DciA-%20and%20DnaC%5C%2FI-helicases%2C%20is%20critical%20in%20vivo.%20Our%20work%20is%20the%20first%20step%20toward%20the%20understanding%20of%20the%20ancestral%20mode%20of%20loading%20of%20bacterial%20replicative%20helicases%20on%20DNA.%20It%20sheds%20light%20on%20the%20strategy%20employed%20by%20phage%20helicase%20loaders%20to%20hijack%20bacterial%20replicative%20helicases%20and%20may%20explain%20the%20recurrent%20domestication%20of%20dnaC%5C%2FI%20through%20evolution%20in%20bacteria.%22%2C%22date%22%3A%222021-06-21%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%2210.1093%5C%2Fnar%5C%2Fgkab463%22%2C%22ISSN%22%3A%220305-1048%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkab463%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222023-05-17T11%3A44%3A31Z%22%7D%7D%2C%7B%22key%22%3A%22AF3N3LWT%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Courtot%20et%20al.%22%2C%22parsedDate%22%3A%222021-05-07%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ECourtot%2C%20L.%2C%20Bournique%2C%20E.%2C%20Maric%2C%20C.%2C%20Guitton-Sert%2C%20L.%2C%20Madrid-Menc%26%23xED%3Ba%2C%20M.%2C%20Pancaldi%2C%20V.%2C%20Cadoret%2C%20J.-C.%2C%20Hoffmann%2C%20J.-S.%2C%20%26amp%3B%20Bergoglio%2C%20V.%20%282021%29.%20Low%20Replicative%20Stress%20Triggers%20Cell-Type%20Specific%20Inheritable%20Advanced%20Replication%20Timing.%20%3Ci%3EInternational%20Journal%20of%20Molecular%20Sciences%3C%5C%2Fi%3E%2C%20%3Ci%3E22%3C%5C%2Fi%3E%289%29%2C%204959.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fijms22094959%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fijms22094959%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Low%20Replicative%20Stress%20Triggers%20Cell-Type%20Specific%20Inheritable%20Advanced%20Replication%20Timing%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lilas%22%2C%22lastName%22%3A%22Courtot%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elodie%22%2C%22lastName%22%3A%22Bournique%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Chrystelle%22%2C%22lastName%22%3A%22Maric%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laure%22%2C%22lastName%22%3A%22Guitton-Sert%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Miguel%22%2C%22lastName%22%3A%22Madrid-Menc%5Cu00eda%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vera%22%2C%22lastName%22%3A%22Pancaldi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Charles%22%2C%22lastName%22%3A%22Cadoret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-S%5Cu00e9bastien%22%2C%22lastName%22%3A%22Hoffmann%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Val%5Cu00e9rie%22%2C%22lastName%22%3A%22Bergoglio%22%7D%5D%2C%22abstractNote%22%3A%22DNA%20replication%20timing%20%28RT%29%2C%20reflecting%20the%20temporal%20order%20of%20origin%20activation%2C%20is%20known%20as%20a%20robust%20and%20conserved%20cell-type%20specific%20process.%20Upon%20low%20replication%20stress%2C%20the%20slowing%20of%20replication%20forks%20induces%20well-documented%20RT%20delays%20associated%20to%20genetic%20instability%2C%20but%20it%20can%20also%20generate%20RT%20advances%20that%20are%20still%20uncharacterized.%20In%20order%20to%20characterize%20these%20advanced%20initiation%20events%2C%20we%20monitored%20the%20whole%20genome%20RT%20from%20six%20independent%20human%20cell%20lines%20treated%20with%20low%20doses%20of%20aphidicolin.%20We%20report%20that%20RT%20advances%20are%20cell-type-specific%20and%20involve%20large%20heterochromatin%20domains.%20Importantly%2C%20we%20found%20that%20some%20major%20late%20to%20early%20RT%20advances%20can%20be%20inherited%20by%20the%20unstressed%20next-cellular%20generation%2C%20which%20is%20a%20unique%20process%20that%20correlates%20with%20enhanced%20chromatin%20accessibility%2C%20as%20well%20as%20modified%20replication%20origin%20landscape%20and%20gene%20expression%20in%20daughter%20cells.%20Collectively%2C%20this%20work%20highlights%20how%20low%20replication%20stress%20may%20impact%20cellular%20identity%20by%20RT%20advances%20events%20at%20a%20subset%20of%20chromosomal%20domains.%22%2C%22date%22%3A%222021-05-07%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.3390%5C%2Fijms22094959%22%2C%22ISSN%22%3A%221422-0067%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A24Z%22%7D%7D%2C%7B%22key%22%3A%222DZICUUN%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Hadjadj%20et%20al.%22%2C%22parsedDate%22%3A%222020-06%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EHadjadj%2C%20D.%2C%20Denecker%2C%20T.%2C%20Gu%26%23xE9%3Brin%2C%20E.%2C%20Kim%2C%20S.-J.%2C%20Fauchereau%2C%20F.%2C%20Baldacci%2C%20G.%2C%20Maric%2C%20C.%2C%20%26amp%3B%20Cadoret%2C%20J.-C.%20%282020%29.%20Efficient%2C%20quick%20and%20easy-to-use%20DNA%20replication%20timing%20analysis%20with%20START-R%20suite.%20%3Ci%3ENAR%20Genomics%20and%20Bioinformatics%3C%5C%2Fi%3E%2C%20%3Ci%3E2%3C%5C%2Fi%3E%282%29%2C%20lqaa045.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnargab%5C%2Flqaa045%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnargab%5C%2Flqaa045%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Efficient%2C%20quick%20and%20easy-to-use%20DNA%20replication%20timing%20analysis%20with%20START-R%20suite%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Djihad%22%2C%22lastName%22%3A%22Hadjadj%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thomas%22%2C%22lastName%22%3A%22Denecker%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eva%22%2C%22lastName%22%3A%22Gu%5Cu00e9rin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Su-Jung%22%2C%22lastName%22%3A%22Kim%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fabien%22%2C%22lastName%22%3A%22Fauchereau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Giuseppe%22%2C%22lastName%22%3A%22Baldacci%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Chrystelle%22%2C%22lastName%22%3A%22Maric%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Charles%22%2C%22lastName%22%3A%22Cadoret%22%7D%5D%2C%22abstractNote%22%3A%22DNA%20replication%20must%20be%20faithful%20and%20follow%20a%20well-defined%20spatiotemporal%20program%20closely%20linked%20to%20transcriptional%20activity%2C%20epigenomic%20marks%2C%20intranuclear%20structures%2C%20mutation%20rate%20and%20cell%20fate%20determination.%20Among%20the%20readouts%20of%20the%20spatiotemporal%20program%20of%20DNA%20replication%2C%20replication%20timing%20analyses%20require%20not%20only%20complex%20and%20time-consuming%20experimental%20procedures%2C%20but%20also%20skills%20in%20bioinformatics.%20We%20developed%20a%20dedicated%20Shiny%20interactive%20web%20application%2C%20the%20START-R%5Cu00a0%28Simple%20Tool%20for%20the%20Analysis%20of%20the%20Replication%20Timing%20based%20on%20R%29%20suite%2C%20which%20analyzes%20DNA%20replication%20timing%20in%20a%20given%20organism%20with%20high-throughput%20data.%20It%20reduces%20the%20time%20required%20for%20generating%20and%20analyzing%20simultaneously%20data%20from%20several%20samples.%20It%20automatically%20detects%20different%20types%20of%20timing%20regions%20and%20identifies%20significant%20differences%20between%20two%20experimental%20conditions%20in%20%5Cu223c15%20min.%20In%20conclusion%2C%20START-R%20suite%20allows%20quick%2C%20efficient%20and%20easier%20analyses%20of%20DNA%20replication%20timing%20for%20all%20organisms.%20This%20novel%20approach%20can%20be%20used%20by%20every%20biologist.%20It%20is%20now%20simpler%20to%20use%20this%20method%20in%20order%20to%20understand%2C%20for%20example%2C%20whether%20%27a%20favorite%20gene%20or%20protein%27%20has%20an%20impact%20on%20replication%20process%20or%2C%20indirectly%2C%20on%20genomic%20organization%20%28as%20Hi-C%20experiments%29%2C%20by%20comparing%20the%20replication%20timing%20profiles%20between%20wild-type%20and%20mutant%20cell%20lines.%22%2C%22date%22%3A%222020-06%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1093%5C%2Fnargab%5C%2Flqaa045%22%2C%22ISSN%22%3A%222631-9268%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A39Z%22%7D%7D%2C%7B%22key%22%3A%22QWTD4EBV%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Hatem%20et%20al.%22%2C%22parsedDate%22%3A%222019-06-01%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EHatem%2C%20E.%2C%20Azzi%2C%20S.%2C%20El%20Banna%2C%20N.%2C%20He%2C%20T.%2C%20Heneman-Masurel%2C%20A.%2C%20Vernis%2C%20L.%2C%20Ba%26%23xEF%3Blle%2C%20D.%2C%20Masson%2C%20V.%2C%20Dingli%2C%20F.%2C%20Loew%2C%20D.%2C%20Azzarone%2C%20B.%2C%20Eid%2C%20P.%2C%20Baldacci%2C%20G.%2C%20%26amp%3B%20Huang%2C%20M.-E.%20%282019%29.%20Auranofin%5C%2FVitamin%20C%3A%20A%20Novel%20Drug%20Combination%20Targeting%20Triple-Negative%20Breast%20Cancer.%20%3Ci%3EJNCI%3A%20Journal%20of%20the%20National%20Cancer%20Institute%3C%5C%2Fi%3E%2C%20%3Ci%3E111%3C%5C%2Fi%3E%286%29%2C%20597%26%23×2013%3B608.%20%3Ca%20class%3D%27zp-ItemURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fjnci%5C%2Fdjy149%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fjnci%5C%2Fdjy149%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Auranofin%5C%2FVitamin%20C%3A%20A%20Novel%20Drug%20Combination%20Targeting%20Triple-Negative%20Breast%20Cancer%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elie%22%2C%22lastName%22%3A%22Hatem%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandy%22%2C%22lastName%22%3A%22Azzi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nadine%22%2C%22lastName%22%3A%22El%20Banna%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Tiantian%22%2C%22lastName%22%3A%22He%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Am%5Cu00e9lie%22%2C%22lastName%22%3A%22Heneman-Masurel%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laurence%22%2C%22lastName%22%3A%22Vernis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Doroth%5Cu00e9e%22%2C%22lastName%22%3A%22Ba%5Cu00eflle%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Masson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Florent%22%2C%22lastName%22%3A%22Dingli%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Damarys%22%2C%22lastName%22%3A%22Loew%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bruno%22%2C%22lastName%22%3A%22Azzarone%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pierre%22%2C%22lastName%22%3A%22Eid%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Giuseppe%22%2C%22lastName%22%3A%22Baldacci%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Meng-Er%22%2C%22lastName%22%3A%22Huang%22%7D%5D%2C%22abstractNote%22%3A%22Cancer%20cells%20from%20different%20origins%20exhibit%20various%20basal%20redox%20statuses%20and%20thus%20respond%20differently%20to%20intrinsic%20or%20extrinsic%20oxidative%20stress.%20These%20intricate%20characteristics%20condition%20the%20success%20of%20redox-based%20anticancer%20therapies%20that%20capitalize%20on%20the%20ability%20of%20reactive%20oxygen%20species%20to%20achieve%20selective%20and%20efficient%20cancer%20cell%20killing.Redox%20biology%20methods%2C%20stable%20isotope%20labeling%20by%20amino%20acids%20in%20cell%20culture%20%28SILAC%29-based%20proteomics%2C%20and%20bioinformatics%20pattern%20comparisons%20were%20used%20to%20decipher%20the%20underlying%20mechanisms%20for%20differential%20response%20of%20lung%20and%20breast%20cancer%20cell%20models%20to%20redox-modulating%20molecule%20auranofin%20%28AUF%29%20and%20to%20combinations%20of%20AUF%20and%20vitamin%20C%20%28VC%29.%20The%20in%20vivo%20effect%20of%20AUF%2C%20VC%2C%20and%20two%20AUF%5C%2FVC%20combinations%20on%20mice%20bearing%20MDA-MB-231%20xenografts%20%28n%5Cu2009%3D%5Cu20095%20mice%20per%20group%29%20was%20also%20evaluated.%20All%20statistical%20tests%20were%20two-sided.AUF%20targeted%20simultaneously%20the%20thioredoxin%20and%20glutathione%20antioxidant%20systems.%20AUF%5C%2FVC%20combinations%20exerted%20a%20synergistic%20and%20hydrogen%20peroxide%20%28H2O2%29-mediated%20cytotoxicity%20toward%20MDA-MB-231%20cells%20and%20other%20breast%20cancer%20cell%20lines.%20The%20anticancer%20potential%20of%20AUF%5C%2FVC%20combinations%20was%20validated%20in%20vivo%20on%20MDA-MB-231%20xenografts%20in%20mice%20without%20notable%20side%20effects.%20On%20day%2014%20of%20treatments%2C%20mean%20%28SD%29%20tumor%20volumes%20for%20the%20vehicle-treated%20control%20group%20and%20the%20two%20AUF%5C%2FVC%20combination%5Cu2013treated%20groups%20%28A%5C%2FV1%20and%20A%5C%2FV2%29%20were%20197.67%20%2824.28%29%20mm3%2C%2015.66%20%2810.90%29%20mm3%2C%20and%2010.23%20%287.30%29mm3%2C%20respectively%3B%20adjusted%20P%20values%20of%20the%20differences%20between%20mean%20tumor%20volumes%20of%20vehicle%20vs%20A%5C%2FV1%20groups%20and%20vehicle%20vs%20A%5C%2FV2%20groups%20were%20both%20less%20than%20.001.%20SILAC%20proteomics%2C%20bioinformatics%20analysis%2C%20and%20functional%20experiments%20linked%20prostaglandin%20reductase%201%20%28PTGR1%29%20expression%20levels%20with%20breast%20cancer%20cell%20sensitivity%20to%20AUF%5C%2FVC%20combinations.The%20combination%20of%20AUF%20and%20VC%2C%20two%20commonly%20available%20drugs%2C%20could%20be%20efficient%20against%20triple-negative%20breast%20cancer%20and%20potentially%20other%20cancers%20with%20similar%20redox%20properties%20and%20PTGR1%20expression%20levels.%20The%20redox-based%20anticancer%20activity%20of%20this%20combination%20and%20the%20discriminatory%20potential%20of%20PTGR1%20expression%20are%20worth%20further%20assessment%20in%20preclinical%20and%20clinical%20studies.%22%2C%22date%22%3A%222019-06-01%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%2210.1093%5C%2Fjnci%5C%2Fdjy149%22%2C%22ISSN%22%3A%220027-8874%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fjnci%5C%2Fdjy149%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222023-05-17T11%3A42%3A15Z%22%7D%7D%2C%7B%22key%22%3A%22YDZZRN3R%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Gialluisi%20et%20al.%22%2C%22parsedDate%22%3A%222019-02-11%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EGialluisi%2C%20A.%2C%20Andlauer%2C%20T.%20F.%20M.%2C%20Mirza-Schreiber%2C%20N.%2C%20Moll%2C%20K.%2C%20Becker%2C%20J.%2C%20Hoffmann%2C%20P.%2C%20Ludwig%2C%20K.%20U.%2C%20Czamara%2C%20D.%2C%20St%20Pourcain%2C%20B.%2C%20Brandler%2C%20W.%2C%20Honbolyg%26%23xF3%3B%2C%20F.%2C%20T%26%23xF3%3Bth%2C%20D.%2C%20Cs%26%23xE9%3Bpe%2C%20V.%2C%20Huguet%2C%20G.%2C%20Morris%2C%20A.%20P.%2C%20Hulslander%2C%20J.%2C%20Willcutt%2C%20E.%20G.%2C%20DeFries%2C%20J.%20C.%2C%20Olson%2C%20R.%20K.%2C%20%26%23×2026%3B%20Schulte-K%26%23xF6%3Brne%2C%20G.%20%282019%29.%20Genome-wide%20association%20scan%20identifies%20new%20variants%20associated%20with%20a%20cognitive%20predictor%20of%20dyslexia.%20%3Ci%3ETranslational%20Psychiatry%3C%5C%2Fi%3E%2C%20%3Ci%3E9%3C%5C%2Fi%3E%281%29%2C%201%26%23×2013%3B15.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41398-019-0402-0%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41398-019-0402-0%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Genome-wide%20association%20scan%20identifies%20new%20variants%20associated%20with%20a%20cognitive%20predictor%20of%20dyslexia%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alessandro%22%2C%22lastName%22%3A%22Gialluisi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Till%20F.%20M.%22%2C%22lastName%22%3A%22Andlauer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nazanin%22%2C%22lastName%22%3A%22Mirza-Schreiber%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kristina%22%2C%22lastName%22%3A%22Moll%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jessica%22%2C%22lastName%22%3A%22Becker%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Per%22%2C%22lastName%22%3A%22Hoffmann%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kerstin%20U.%22%2C%22lastName%22%3A%22Ludwig%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Darina%22%2C%22lastName%22%3A%22Czamara%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Beate%22%2C%22lastName%22%3A%22St%20Pourcain%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22William%22%2C%22lastName%22%3A%22Brandler%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ferenc%22%2C%22lastName%22%3A%22Honbolyg%5Cu00f3%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22D%5Cu00e9nes%22%2C%22lastName%22%3A%22T%5Cu00f3th%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Val%5Cu00e9ria%22%2C%22lastName%22%3A%22Cs%5Cu00e9pe%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Guillaume%22%2C%22lastName%22%3A%22Huguet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Andrew%20P.%22%2C%22lastName%22%3A%22Morris%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jacqueline%22%2C%22lastName%22%3A%22Hulslander%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Erik%20G.%22%2C%22lastName%22%3A%22Willcutt%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22John%20C.%22%2C%22lastName%22%3A%22DeFries%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Richard%20K.%22%2C%22lastName%22%3A%22Olson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Shelley%20D.%22%2C%22lastName%22%3A%22Smith%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bruce%20F.%22%2C%22lastName%22%3A%22Pennington%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anniek%22%2C%22lastName%22%3A%22Vaessen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Urs%22%2C%22lastName%22%3A%22Maurer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Heikki%22%2C%22lastName%22%3A%22Lyytinen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Myriam%22%2C%22lastName%22%3A%22Peyrard-Janvid%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Paavo%20H.%20T.%22%2C%22lastName%22%3A%22Lepp%5Cu00e4nen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Daniel%22%2C%22lastName%22%3A%22Brandeis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Milene%22%2C%22lastName%22%3A%22Bonte%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22John%20F.%22%2C%22lastName%22%3A%22Stein%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Joel%20B.%22%2C%22lastName%22%3A%22Talcott%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fabien%22%2C%22lastName%22%3A%22Fauchereau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Arndt%22%2C%22lastName%22%3A%22Wilcke%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Clyde%22%2C%22lastName%22%3A%22Francks%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thomas%22%2C%22lastName%22%3A%22Bourgeron%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anthony%20P.%22%2C%22lastName%22%3A%22Monaco%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Franck%22%2C%22lastName%22%3A%22Ramus%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Karin%22%2C%22lastName%22%3A%22Landerl%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Juha%22%2C%22lastName%22%3A%22Kere%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thomas%20S.%22%2C%22lastName%22%3A%22Scerri%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Silvia%22%2C%22lastName%22%3A%22Paracchini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Simon%20E.%22%2C%22lastName%22%3A%22Fisher%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Johannes%22%2C%22lastName%22%3A%22Schumacher%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Markus%20M.%22%2C%22lastName%22%3A%22N%5Cu00f6then%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bertram%22%2C%22lastName%22%3A%22M%5Cu00fcller-Myhsok%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gerd%22%2C%22lastName%22%3A%22Schulte-K%5Cu00f6rne%22%7D%5D%2C%22abstractNote%22%3A%22Developmental%20dyslexia%20%28DD%29%20is%20one%20of%20the%20most%20prevalent%20learning%20disorders%2C%20with%20high%20impact%20on%20school%20and%20psychosocial%20development%20and%20high%20comorbidity%20with%20conditions%20like%20attention-deficit%20hyperactivity%20disorder%20%28ADHD%29%2C%20depression%2C%20and%20anxiety.%20DD%20is%20characterized%20by%20deficits%20in%20different%20cognitive%20skills%2C%20including%20word%20reading%2C%20spelling%2C%20rapid%20naming%2C%20and%20phonology.%20To%20investigate%20the%20genetic%20basis%20of%20DD%2C%20we%20conducted%20a%20genome-wide%20association%20study%20%28GWAS%29%20of%20these%20skills%20within%20one%20of%20the%20largest%20studies%20available%2C%20including%20nine%20cohorts%20of%20reading-impaired%20and%20typically%20developing%20children%20of%20European%20ancestry%20%28N%5Cu2009%3D%5Cu20092562%5Cu20133468%29.%20We%20observed%20a%20genome-wide%20significant%20effect%20%28p%5Cu2009%3C%5Cu20091%5Cu2009%5Cu00d7%5Cu200910%5Cu22128%29%20on%20rapid%20automatized%20naming%20of%20letters%20%28RANlet%29%20for%20variants%20on%2018q12.2%2C%20within%20MIR924HG%20%28micro-RNA%20924%20host%20gene%3B%20rs17663182%20p%5Cu2009%3D%5Cu20094.73%5Cu2009%5Cu00d7%5Cu200910%5Cu22129%29%2C%20and%20a%20suggestive%20association%20on%208q12.3%20within%20NKAIN3%20%28encoding%20a%20cation%20transporter%3B%20rs16928927%2C%20p%5Cu2009%3D%5Cu20092.25%5Cu2009%5Cu00d7%5Cu200910%5Cu22128%29.%20rs17663182%20%2818q12.2%29%20also%20showed%20genome-wide%20significant%20multivariate%20associations%20with%20RAN%20measures%20%28p%5Cu2009%3D%5Cu20091.15%5Cu2009%5Cu00d7%5Cu200910%5Cu22128%29%20and%20with%20all%20the%20cognitive%20traits%20tested%20%28p%5Cu2009%3D%5Cu20093.07%5Cu2009%5Cu00d7%5Cu200910%5Cu22128%29%2C%20suggesting%20%28relational%29%20pleiotropic%20effects%20of%20this%20variant.%20A%20polygenic%20risk%20score%20%28PRS%29%20analysis%20revealed%20significant%20genetic%20overlaps%20of%20some%20of%20the%20DD-related%20traits%20with%20educational%20attainment%20%28EDUyears%29%20and%20ADHD.%20Reading%20and%20spelling%20abilities%20were%20positively%20associated%20with%20EDUyears%20%28p%5Cu2009~%5Cu2009%5B10%5Cu22125%5Cu201310%5Cu22127%5D%29%20and%20negatively%20associated%20with%20ADHD%20PRS%20%28p%5Cu2009~%5Cu2009%5B10%5Cu22128%5Cu221210%5Cu221217%5D%29.%20This%20corroborates%20a%20long-standing%20hypothesis%20on%20the%20partly%20shared%20genetic%20etiology%20of%20DD%20and%20ADHD%2C%20at%20the%20genome-wide%20level.%20Our%20findings%20suggest%20new%20candidate%20DD%20susceptibility%20genes%20and%20provide%20new%20insights%20into%20the%20genetics%20of%20dyslexia%20and%20its%20comorbities.%22%2C%22date%22%3A%222019-02-11%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41398-019-0402-0%22%2C%22ISSN%22%3A%222158-3188%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.nature.com%5C%2Farticles%5C%2Fs41398-019-0402-0%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222023-05-17T11%3A42%3A40Z%22%7D%7D%2C%7B%22key%22%3A%22YJVAV5IU%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Maes%20et%20al.%22%2C%22parsedDate%22%3A%222018-08%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EMaes%2C%20P.%2C%20Alkhovsky%2C%20S.%20V.%2C%20B%26%23xE0%3Bo%2C%20Y.%2C%20Beer%2C%20M.%2C%20Birkhead%2C%20M.%2C%20Briese%2C%20T.%2C%20Buchmeier%2C%20M.%20J.%2C%20Calisher%2C%20C.%20H.%2C%20Charrel%2C%20R.%20N.%2C%20Choi%2C%20I.%20R.%2C%20Clegg%2C%20C.%20S.%2C%20de%20la%20Torre%2C%20J.%20C.%2C%20Delwart%2C%20E.%2C%20DeRisi%2C%20J.%20L.%2C%20Di%20Bello%2C%20P.%20L.%2C%20Di%20Serio%2C%20F.%2C%20Digiaro%2C%20M.%2C%20Dolja%2C%20V.%20V.%2C%20Drosten%2C%20C.%2C%20%26%23×2026%3B%20Kuhn%2C%20J.%20H.%20%282018%29.%20Taxonomy%20of%20the%20family%20Arenaviridae%20and%20the%20order%20Bunyavirales%3A%20update%202018.%20%3Ci%3EArchives%20of%20Virology%3C%5C%2Fi%3E%2C%20%3Ci%3E163%3C%5C%2Fi%3E%288%29%2C%202295%26%23×2013%3B2310.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00705-018-3843-5%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00705-018-3843-5%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Taxonomy%20of%20the%20family%20Arenaviridae%20and%20the%20order%20Bunyavirales%3A%20update%202018%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Piet%22%2C%22lastName%22%3A%22Maes%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sergey%20V.%22%2C%22lastName%22%3A%22Alkhovsky%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Y%5Cu012bm%5Cu00edng%22%2C%22lastName%22%3A%22B%5Cu00e0o%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Martin%22%2C%22lastName%22%3A%22Beer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Monica%22%2C%22lastName%22%3A%22Birkhead%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thomas%22%2C%22lastName%22%3A%22Briese%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michael%20J.%22%2C%22lastName%22%3A%22Buchmeier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Charles%20H.%22%2C%22lastName%22%3A%22Calisher%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R%5Cu00e9mi%20N.%22%2C%22lastName%22%3A%22Charrel%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Il%20Ryong%22%2C%22lastName%22%3A%22Choi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Christopher%20S.%22%2C%22lastName%22%3A%22Clegg%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Juan%20Carlos%22%2C%22lastName%22%3A%22de%20la%20Torre%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eric%22%2C%22lastName%22%3A%22Delwart%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Joseph%20L.%22%2C%22lastName%22%3A%22DeRisi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Patrick%20L.%22%2C%22lastName%22%3A%22Di%20Bello%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Francesco%22%2C%22lastName%22%3A%22Di%20Serio%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michele%22%2C%22lastName%22%3A%22Digiaro%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Valerian%20V.%22%2C%22lastName%22%3A%22Dolja%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Christian%22%2C%22lastName%22%3A%22Drosten%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Tobiasz%20Z.%22%2C%22lastName%22%3A%22Druciarek%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jiang%22%2C%22lastName%22%3A%22Du%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hideki%22%2C%22lastName%22%3A%22Ebihara%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Toufic%22%2C%22lastName%22%3A%22Elbeaino%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rose%20C.%22%2C%22lastName%22%3A%22Gergerich%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Amethyst%20N.%22%2C%22lastName%22%3A%22Gillis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Paul%20J.%22%2C%22lastName%22%3A%22Gonzalez%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anne-Lise%22%2C%22lastName%22%3A%22Haenni%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jussi%22%2C%22lastName%22%3A%22Hepojoki%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Udo%22%2C%22lastName%22%3A%22Hetzel%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thi%5Cu1ec7n%22%2C%22lastName%22%3A%22H%5Cu1ed3%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22N%5Cu00ed%22%2C%22lastName%22%3A%22H%5Cu00f3ng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rakesh%20K.%22%2C%22lastName%22%3A%22Jain%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Petrus%22%2C%22lastName%22%3A%22Jansen%20van%20Vuren%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Qi%22%2C%22lastName%22%3A%22Jin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Miranda%20Gilda%22%2C%22lastName%22%3A%22Jonson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandra%22%2C%22lastName%22%3A%22Junglen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Karen%20E.%22%2C%22lastName%22%3A%22Keller%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alan%22%2C%22lastName%22%3A%22Kemp%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anja%22%2C%22lastName%22%3A%22Kipar%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nikola%20O.%22%2C%22lastName%22%3A%22Kondov%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eugene%20V.%22%2C%22lastName%22%3A%22Koonin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Richard%22%2C%22lastName%22%3A%22Kormelink%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yegor%22%2C%22lastName%22%3A%22Korzyukov%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mart%22%2C%22lastName%22%3A%22Krupovic%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Amy%20J.%22%2C%22lastName%22%3A%22Lambert%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alma%20G.%22%2C%22lastName%22%3A%22Laney%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Matthew%22%2C%22lastName%22%3A%22LeBreton%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Igor%20S.%22%2C%22lastName%22%3A%22Lukashevich%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marco%22%2C%22lastName%22%3A%22Marklewitz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Wanda%22%2C%22lastName%22%3A%22Markotter%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Giovanni%20P.%22%2C%22lastName%22%3A%22Martelli%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Robert%20R.%22%2C%22lastName%22%3A%22Martin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicole%22%2C%22lastName%22%3A%22Mielke-Ehret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hans-Peter%22%2C%22lastName%22%3A%22M%5Cu00fchlbach%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Beatriz%22%2C%22lastName%22%3A%22Navarro%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Terry%20Fei%20Fan%22%2C%22lastName%22%3A%22Ng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M%5Cu00e1rcio%20Roberto%20Teixeira%22%2C%22lastName%22%3A%22Nunes%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gustavo%22%2C%22lastName%22%3A%22Palacios%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Janusz%20T.%22%2C%22lastName%22%3A%22Paw%5Cu0119ska%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Clarence%20J.%22%2C%22lastName%22%3A%22Peters%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alexander%22%2C%22lastName%22%3A%22Plyusnin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sheli%20R.%22%2C%22lastName%22%3A%22Radoshitzky%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22V%5Cu00edctor%22%2C%22lastName%22%3A%22Romanowski%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pertteli%22%2C%22lastName%22%3A%22Salmenper%5Cu00e4%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Maria%20S.%22%2C%22lastName%22%3A%22Salvato%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22H%5Cu00e9l%5Cu00e8ne%22%2C%22lastName%22%3A%22Sanfa%5Cu00e7on%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Takahide%22%2C%22lastName%22%3A%22Sasaya%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Connie%22%2C%22lastName%22%3A%22Schmaljohn%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bradley%20S.%22%2C%22lastName%22%3A%22Schneider%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yukio%22%2C%22lastName%22%3A%22Shirako%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Stuart%22%2C%22lastName%22%3A%22Siddell%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Tarja%20A.%22%2C%22lastName%22%3A%22Sironen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mark%20D.%22%2C%22lastName%22%3A%22Stenglein%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nadia%22%2C%22lastName%22%3A%22Storm%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Harikishan%22%2C%22lastName%22%3A%22Sudini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Robert%20B.%22%2C%22lastName%22%3A%22Tesh%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ioannis%20E.%22%2C%22lastName%22%3A%22Tzanetakis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mangala%22%2C%22lastName%22%3A%22Uppala%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Olli%22%2C%22lastName%22%3A%22Vapalahti%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nikos%22%2C%22lastName%22%3A%22Vasilakis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%20J.%22%2C%22lastName%22%3A%22Walker%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gu%5Cu00f3p%5Cu00edng%22%2C%22lastName%22%3A%22W%5Cu00e1ng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L%5Cu00ecp%5Cu00edng%22%2C%22lastName%22%3A%22W%5Cu00e1ng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Y%5Cu00e0nxi%5Cu0103ng%22%2C%22lastName%22%3A%22W%5Cu00e1ng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22T%5Cu00e0iy%5Cu00fan%22%2C%22lastName%22%3A%22W%5Cu00e8i%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michael%20R.%22%2C%22lastName%22%3A%22Wiley%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yuri%20I.%22%2C%22lastName%22%3A%22Wolf%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nathan%20D.%22%2C%22lastName%22%3A%22Wolfe%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Zh%5Cu00ecqi%5Cu00e1ng%22%2C%22lastName%22%3A%22W%5Cu00fa%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22W%5Cu00e9nx%5Cu00ecng%22%2C%22lastName%22%3A%22X%5Cu00fa%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Li%22%2C%22lastName%22%3A%22Yang%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Zu%5Cu00f2k%5Cu016bn%22%2C%22lastName%22%3A%22Y%5Cu0101ng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Shyi-Dong%22%2C%22lastName%22%3A%22Yeh%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Y%5Cu01d2ng-Zh%5Cu00e8n%22%2C%22lastName%22%3A%22Zh%5Cu0101ng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Y%5Cu00e0zh%5Cu014du%22%2C%22lastName%22%3A%22Zh%5Cu00e8ng%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Xueping%22%2C%22lastName%22%3A%22Zhou%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ch%5Cu00e9nx%5Cu012b%22%2C%22lastName%22%3A%22Zh%5Cu016b%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Florian%22%2C%22lastName%22%3A%22Zirkel%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jens%20H.%22%2C%22lastName%22%3A%22Kuhn%22%7D%5D%2C%22abstractNote%22%3A%22In%202018%2C%20the%20family%20Arenaviridae%20was%20expanded%20by%20inclusion%20of%201%20new%20genus%20and%205%20novel%20species.%20At%20the%20same%20time%2C%20the%20recently%20established%20order%20Bunyavirales%20was%20expanded%20by%203%20species.%20This%20article%20presents%20the%20updated%20taxonomy%20of%20the%20family%20Arenaviridae%20and%20the%20order%20Bunyavirales%20as%20now%20accepted%20by%20the%20International%20Committee%20on%20Taxonomy%20of%20Viruses%20%28ICTV%29%20and%20summarizes%20additional%20taxonomic%20proposals%20that%20may%20affect%20the%20order%20in%20the%20near%20future.%22%2C%22date%22%3A%222018-08%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1007%5C%2Fs00705-018-3843-5%22%2C%22ISSN%22%3A%221432-8798%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A49Z%22%7D%7D%2C%7B%22key%22%3A%22TJ2JLYZN%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Almeida%20et%20al.%22%2C%22parsedDate%22%3A%222018-04-23%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EAlmeida%2C%20R.%2C%20Fern%26%23xE1%3Bndez-Justel%2C%20J.%20M.%2C%20Santa-Mar%26%23xED%3Ba%2C%20C.%2C%20Cadoret%2C%20J.-C.%2C%20Cano-Aroca%2C%20L.%2C%20Lombra%26%23xF1%3Ba%2C%20R.%2C%20Herranz%2C%20G.%2C%20Agresti%2C%20A.%2C%20%26amp%3B%20G%26%23xF3%3Bmez%2C%20M.%20%282018%29.%20Chromatin%20conformation%20regulates%20the%20coordination%20between%20DNA%20replication%20and%20transcription.%20%3Ci%3ENature%20Communications%3C%5C%2Fi%3E%2C%20%3Ci%3E9%3C%5C%2Fi%3E%281%29%2C%201590.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41467-018-03539-8%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41467-018-03539-8%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Chromatin%20conformation%20regulates%20the%20coordination%20between%20DNA%20replication%20and%20transcription%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ricardo%22%2C%22lastName%22%3A%22Almeida%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jos%5Cu00e9%20Miguel%22%2C%22lastName%22%3A%22Fern%5Cu00e1ndez-Justel%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cristina%22%2C%22lastName%22%3A%22Santa-Mar%5Cu00eda%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Charles%22%2C%22lastName%22%3A%22Cadoret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laura%22%2C%22lastName%22%3A%22Cano-Aroca%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rodrigo%22%2C%22lastName%22%3A%22Lombra%5Cu00f1a%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gonzalo%22%2C%22lastName%22%3A%22Herranz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alessandra%22%2C%22lastName%22%3A%22Agresti%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mar%5Cu00eda%22%2C%22lastName%22%3A%22G%5Cu00f3mez%22%7D%5D%2C%22abstractNote%22%3A%22Chromatin%20is%20the%20template%20for%20the%20basic%20processes%20of%20replication%20and%20transcription%2C%20making%20the%20maintenance%20of%20chromosomal%20integrity%20critical%20for%20cell%20viability.%20To%20elucidate%20how%20dividing%20cells%20respond%20to%20alterations%20in%20chromatin%20structure%2C%20here%20we%20analyse%20the%20replication%20programme%20of%20primary%20cells%20with%20altered%20chromatin%20configuration%20caused%20by%20the%20genetic%20ablation%20of%20the%20HMGB1%20gene%2C%20or%20three%20histone%20H1%20genes.%20We%20find%20that%20loss%20of%20chromatin%20compaction%20in%20H1-depleted%20cells%20triggers%20the%20accumulation%20of%20stalled%20forks%20and%20DNA%20damage%20as%20a%20consequence%20of%20transcription-replication%20conflicts.%20In%20contrast%2C%20reductions%20in%20nucleosome%20occupancy%20due%20to%20the%20lack%20of%20HMGB1%20cause%20faster%20fork%20progression%20without%20impacting%20the%20initiation%20landscape%20or%20fork%20stability.%20Thus%2C%20perturbations%20in%20chromatin%20integrity%20elicit%20a%20range%20of%20responses%20in%20the%20dynamics%20of%20DNA%20replication%20and%20transcription%2C%20with%20different%20consequences%20on%20replicative%20stress.%20These%20findings%20have%20broad%20implications%20for%20our%20understanding%20of%20how%20defects%20in%20chromatin%20structure%20contribute%20to%20genomic%20instability.%22%2C%22date%22%3A%222018-04-23%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41467-018-03539-8%22%2C%22ISSN%22%3A%222041-1723%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A49Z%22%7D%7D%2C%7B%22key%22%3A%22MH7452I2%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Hadjadj%20et%20al.%22%2C%22parsedDate%22%3A%222017-12-26%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EHadjadj%2C%20D.%2C%20Kim%2C%20S.-J.%2C%20Denecker%2C%20T.%2C%20Ben%20Driss%2C%20L.%2C%20Cadoret%2C%20J.-C.%2C%20Maric%2C%20C.%2C%20Baldacci%2C%20G.%2C%20%26amp%3B%20Fauchereau%2C%20F.%20%282017%29.%20A%20hypothesis-driven%20approach%20identifies%20CDK4%20and%20CDK6%20inhibitors%20as%20candidate%20drugs%20for%20treatments%20of%20adrenocortical%20carcinomas.%20%3Ci%3EAging%3C%5C%2Fi%3E%2C%20%3Ci%3E9%3C%5C%2Fi%3E%2812%29%2C%202695%26%23×2013%3B2716.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.18632%5C%2Faging.101356%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.18632%5C%2Faging.101356%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20hypothesis-driven%20approach%20identifies%20CDK4%20and%20CDK6%20inhibitors%20as%20candidate%20drugs%20for%20treatments%20of%20adrenocortical%20carcinomas%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Djihad%22%2C%22lastName%22%3A%22Hadjadj%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Su-Jung%22%2C%22lastName%22%3A%22Kim%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thomas%22%2C%22lastName%22%3A%22Denecker%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laura%22%2C%22lastName%22%3A%22Ben%20Driss%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Charles%22%2C%22lastName%22%3A%22Cadoret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Chrystelle%22%2C%22lastName%22%3A%22Maric%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Giuseppe%22%2C%22lastName%22%3A%22Baldacci%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fabien%22%2C%22lastName%22%3A%22Fauchereau%22%7D%5D%2C%22abstractNote%22%3A%22High%20proliferation%20rate%20and%20high%20mutation%20density%20are%20both%20indicators%20of%20poor%20prognosis%20in%20adrenocortical%20carcinomas.%20We%20performed%20a%20hypothesis-driven%20association%20study%20between%20clinical%20features%20in%20adrenocortical%20carcinomas%20and%20the%20expression%20levels%20of%20136%20genes%20involved%20in%20DNA%20metabolism%20and%20G1%5C%2FS%20phase%20transition.%20In%2079%20samples%20downloaded%20from%20The%20Cancer%20Genome%20Atlas%20portal%2C%20high%20Cyclin%20Dependent%20Kinase%206%20%28CDK6%29%20mRNA%20levels%20gave%20the%20most%20significant%20association%20with%20shorter%20time%20to%20relapse%20and%20poorer%20survival%20of%20patients.%20A%20hierarchical%20clustering%20approach%20assembled%20most%20tumors%20with%20high%20levels%20of%20CDK6%20mRNA%20into%20one%20group.%20These%20tumors%20tend%20to%20cumulate%20mutations%20activating%20the%20Wnt%5C%2F%5Cu03b2-catenin%20pathway%20and%20show%20reduced%20MIR506%20expression.%20Actually%2C%20the%20level%20of%20MIR506%20RNA%20is%20inversely%20correlated%20with%20the%20levels%20of%20both%20CDK6%20and%20CTNNB1%20%28encoding%20%5Cu03b2-catenin%29.%20Together%20these%20results%20indicate%20that%20high%20CDK6%20expression%20is%20found%20in%20aggressive%20tumors%20with%20activated%20Wnt%5C%2F%5Cu03b2-catenin%20pathway.%20Thus%20we%20tested%20the%20impact%20of%20Food%20and%20Drug%20Administration-approved%20CDK4%20and%20CDK6%20inhibitors%2C%20namely%20palbociclib%20and%20ribociclib%2C%20on%20SW-13%20and%20NCI-H295R%20cells.%20While%20both%20drugs%20reduced%20viability%20and%20induced%20senescence%20in%20SW-13%20cells%2C%20only%20palbociclib%20was%20effective%20on%20the%20retinoblastoma%20protein%20%28pRB%29-negative%20NCI-H295R%20cells%2C%20by%20inducing%20apoptosis.%20In%20NCI-H295R%20cells%2C%20palbociclib%20induced%20an%20increase%20of%20the%20active%20form%20of%20Glycogen%20Synthase%20Kinase%203%5Cu03b2%20%28GSK3%5Cu03b2%29%20responsible%20for%20the%20reduced%20amount%20of%20active%20%5Cu03b2-catenin%2C%20and%20altered%20the%20amount%20of%20AXIN2%20mRNA.%20Taken%20together%2C%20these%20data%20underline%20the%20impact%20of%20CDK4%20and%20CDK6%20inhibitors%20in%20treating%20adrenocortical%20carcinomas.%22%2C%22date%22%3A%222017-12-26%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.18632%5C%2Faging.101356%22%2C%22ISSN%22%3A%221945-4589%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A28%3A03Z%22%7D%7D%2C%7B%22key%22%3A%22YRC2X2TA%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Brustel%20et%20al.%22%2C%22parsedDate%22%3A%222017-09-15%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EBrustel%2C%20J.%2C%20Kirstein%2C%20N.%2C%20Izard%2C%20F.%2C%20Grimaud%2C%20C.%2C%20Prorok%2C%20P.%2C%20Cayrou%2C%20C.%2C%20Schotta%2C%20G.%2C%20Abdelsamie%2C%20A.%20F.%2C%20D%26%23xE9%3Bjardin%2C%20J.%2C%20M%26%23xE9%3Bchali%2C%20M.%2C%20Baldacci%2C%20G.%2C%20Sardet%2C%20C.%2C%20Cadoret%2C%20J.-C.%2C%20Schepers%2C%20A.%2C%20%26amp%3B%20Julien%2C%20E.%20%282017%29.%20Histone%20H4K20%20tri-methylation%20at%20late-firing%20origins%20ensures%20timely%20heterochromatin%20replication.%20%3Ci%3EThe%20EMBO%20Journal%3C%5C%2Fi%3E%2C%20%3Ci%3E36%3C%5C%2Fi%3E%2818%29%2C%202726%26%23×2013%3B2741.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.15252%5C%2Fembj.201796541%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.15252%5C%2Fembj.201796541%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Histone%20H4K20%20tri-methylation%20at%20late-firing%20origins%20ensures%20timely%20heterochromatin%20replication%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julien%22%2C%22lastName%22%3A%22Brustel%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nina%22%2C%22lastName%22%3A%22Kirstein%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fanny%22%2C%22lastName%22%3A%22Izard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Charlotte%22%2C%22lastName%22%3A%22Grimaud%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Paulina%22%2C%22lastName%22%3A%22Prorok%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Christelle%22%2C%22lastName%22%3A%22Cayrou%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gunnar%22%2C%22lastName%22%3A%22Schotta%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alhassan%20F.%22%2C%22lastName%22%3A%22Abdelsamie%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22J%5Cu00e9r%5Cu00f4me%22%2C%22lastName%22%3A%22D%5Cu00e9jardin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marcel%22%2C%22lastName%22%3A%22M%5Cu00e9chali%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Giuseppe%22%2C%22lastName%22%3A%22Baldacci%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Claude%22%2C%22lastName%22%3A%22Sardet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Charles%22%2C%22lastName%22%3A%22Cadoret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Aloys%22%2C%22lastName%22%3A%22Schepers%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eric%22%2C%22lastName%22%3A%22Julien%22%7D%5D%2C%22abstractNote%22%3A%22Among%20other%20targets%2C%20the%20protein%20lysine%20methyltransferase%20PR-Set7%20induces%20histone%20H4%20lysine%2020%20monomethylation%20%28H4K20me1%29%2C%20which%20is%20the%20substrate%20for%20further%20methylation%20by%20the%20Suv4-20h%20methyltransferase.%20Although%20these%20enzymes%20have%20been%20implicated%20in%20control%20of%20replication%20origins%2C%20the%20specific%20contribution%20of%20H4K20%20methylation%20to%20DNA%20replication%20remains%20unclear.%20Here%2C%20we%20show%20that%20H4K20%20mutation%20in%20mammalian%20cells%2C%20unlike%20in%20Drosophila%2C%20partially%20impairs%20S-phase%20progression%20and%20protects%20from%20DNA%20re-replication%20induced%20by%20stabilization%20of%20PR-Set7.%20Using%20Epstein-Barr%20virus-derived%20episomes%2C%20we%20further%20demonstrate%20that%20conversion%20of%20H4K20me1%20to%20higher%20H4K20me2%5C%2F3%20states%20by%20Suv4-20h%20is%20not%20sufficient%20to%20define%20an%20efficient%20origin%20per%20se%2C%20but%20rather%20serves%20as%20an%20enhancer%20for%20MCM2-7%20helicase%20loading%20and%20replication%20activation%20at%20defined%20origins.%20Consistent%20with%20this%2C%20we%20find%20that%20Suv4-20h-mediated%20H4K20%20tri-methylation%20%28H4K20me3%29%20is%20required%20to%20sustain%20the%20licensing%20and%20activity%20of%20a%20subset%20of%20ORCA%5C%2FLRWD1-associated%20origins%2C%20which%20ensure%20proper%20replication%20timing%20of%20late-replicating%20heterochromatin%20domains.%20Altogether%2C%20these%20results%20reveal%20Suv4-20h-mediated%20H4K20%20tri-methylation%20as%20a%20critical%20determinant%20in%20the%20selection%20of%20active%20replication%20initiation%20sites%20in%20heterochromatin%20regions%20of%20mammalian%20genomes.%22%2C%22date%22%3A%222017-09-15%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.15252%5C%2Fembj.201796541%22%2C%22ISSN%22%3A%221460-2075%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A28%3A03Z%22%7D%7D%2C%7B%22key%22%3A%22W34EHUA4%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Pagan%20et%20al.%22%2C%22parsedDate%22%3A%222017-05-18%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EPagan%2C%20C.%2C%20Goubran-Botros%2C%20H.%2C%20Delorme%2C%20R.%2C%20Benabou%2C%20M.%2C%20Lemi%26%23xE8%3Bre%2C%20N.%2C%20Murray%2C%20K.%2C%20Amsellem%2C%20F.%2C%20Callebert%2C%20J.%2C%20Chaste%2C%20P.%2C%20Jamain%2C%20S.%2C%20Fauchereau%2C%20F.%2C%20Huguet%2C%20G.%2C%20Maronde%2C%20E.%2C%20Leboyer%2C%20M.%2C%20Launay%2C%20J.-M.%2C%20%26amp%3B%20Bourgeron%2C%20T.%20%282017%29.%20Disruption%20of%20melatonin%20synthesis%20is%20associated%20with%20impaired%2014-3-3%20and%20miR-451%20levels%20in%20patients%20with%20autism%20spectrum%20disorders.%20%3Ci%3EScientific%20Reports%3C%5C%2Fi%3E%2C%20%3Ci%3E7%3C%5C%2Fi%3E%281%29%2C%202096.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41598-017-02152-x%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41598-017-02152-x%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Disruption%20of%20melatonin%20synthesis%20is%20associated%20with%20impaired%2014-3-3%20and%20miR-451%20levels%20in%20patients%20with%20autism%20spectrum%20disorders%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C%5Cu00e9cile%22%2C%22lastName%22%3A%22Pagan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hany%22%2C%22lastName%22%3A%22Goubran-Botros%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Richard%22%2C%22lastName%22%3A%22Delorme%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marion%22%2C%22lastName%22%3A%22Benabou%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nathalie%22%2C%22lastName%22%3A%22Lemi%5Cu00e8re%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kerren%22%2C%22lastName%22%3A%22Murray%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fr%5Cu00e9d%5Cu00e9rique%22%2C%22lastName%22%3A%22Amsellem%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jacques%22%2C%22lastName%22%3A%22Callebert%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pauline%22%2C%22lastName%22%3A%22Chaste%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phane%22%2C%22lastName%22%3A%22Jamain%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fabien%22%2C%22lastName%22%3A%22Fauchereau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Guillaume%22%2C%22lastName%22%3A%22Huguet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Erik%22%2C%22lastName%22%3A%22Maronde%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marion%22%2C%22lastName%22%3A%22Leboyer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Marie%22%2C%22lastName%22%3A%22Launay%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thomas%22%2C%22lastName%22%3A%22Bourgeron%22%7D%5D%2C%22abstractNote%22%3A%22Autism%20spectrum%20disorders%20%28ASD%29%20are%20characterized%20by%20a%20wide%20genetic%20and%20clinical%20heterogeneity.%20However%2C%20some%20biochemical%20impairments%2C%20including%20decreased%20melatonin%20%28crucial%20for%20circadian%20regulation%29%20and%20elevated%20platelet%20N-acetylserotonin%20%28the%20precursor%20of%20melatonin%29%20have%20been%20reported%20as%20very%20frequent%20features%20in%20individuals%20with%20ASD.%20To%20address%20the%20mechanisms%20of%20these%20dysfunctions%2C%20we%20investigated%20melatonin%20synthesis%20in%20post-mortem%20pineal%20glands%20-%20the%20main%20source%20of%20melatonin%20%289%20patients%20and%2022%20controls%29%20-%20and%20gut%20samples%20-%20the%20main%20source%20of%20serotonin%20%2811%20patients%20and%2013%20controls%29%2C%20and%20in%20blood%20platelets%20from%20239%20individuals%20with%20ASD%2C%20their%20first-degree%20relatives%20and%20278%20controls.%20Our%20results%20elucidate%20the%20enzymatic%20mechanism%20for%20melatonin%20deficit%20in%20ASD%2C%20involving%20a%20reduction%20of%20both%20enzyme%20activities%20contributing%20to%20melatonin%20synthesis%20%28AANAT%20and%20ASMT%29%2C%20observed%20in%20the%20pineal%20gland%20as%20well%20as%20in%20gut%20and%20platelets%20of%20patients.%20Further%20investigations%20suggest%20new%2C%20post-translational%20%28reduced%20levels%20of%2014-3-3%20proteins%20which%20regulate%20AANAT%20and%20ASMT%20activities%29%20and%20post-transcriptional%20%28increased%20levels%20of%20miR-451%2C%20targeting%2014-3-3%5Cu03b6%29%20mechanisms%20to%20these%20impairments.%20This%20study%20thus%20gives%20insights%20into%20the%20pathophysiological%20pathways%20involved%20in%20ASD.%22%2C%22date%22%3A%222017-05-18%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41598-017-02152-x%22%2C%22ISSN%22%3A%222045-2322%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.nature.com%5C%2Farticles%5C%2Fs41598-017-02152-x%22%2C%22collections%22%3A%5B%22WM7T2BK4%22%5D%2C%22dateModified%22%3A%222023-05-17T11%3A43%3A16Z%22%7D%7D%5D%7D
Göder, A., Maric, C. A., Rainey, M. D., O’Connor, A., Cazzaniga, C., Shamavu, D., Cadoret, J.-C., & Santocanale, C. (2024). DBF4, not DRF1, is the crucial regulator of CDC7 kinase at replication forks.
Journal of Cell Biology,
223(8), e202402144.
https://doi.org/10.1083/jcb.202402144
Lebdy, R., Canut, M., Patouillard, J., Cadoret, J.-C., Letessier, A., Ammar, J., Basbous, J., Urbach, S., Miotto, B., Constantinou, A., Abou Merhi, R., & Ribeyre, C. (2023). The nucleolar protein GNL3 prevents resection of stalled replication forks.
EMBO Reports,
n/a(n/a), e57585.
https://doi.org/10.15252/embr.202357585
Kim, S.-J., Maric, C., Briu, L.-M., Fauchereau, F., Baldacci, G., Debatisse, M., Koundrioukoff, S., & Cadoret, J.-C. (2023). Firing of Replication Origins Is Disturbed by a CDK4/6 Inhibitor in a pRb-Independent Manner.
International Journal of Molecular Sciences,
24(13), 10629.
https://doi.org/10.3390/ijms241310629
Adam, Y., Brezellec, P., Espinosa, E., Besombes, A., Naquin, D., Paly, E., Possoz, C., van Dijk, E., Barre, F.-X., & Ferat, J.-L. (2022). Plesiomonas shigelloides, an Atypical Enterobacterales with a Vibrio-Related Secondary Chromosome.
Genome Biology and Evolution,
14(2), evac011.
https://doi.org/10.1093/gbe/evac011
Ben Yamin, B., Ahmed-Seghir, S., Tomida, J., Despras, E., Pouvelle, C., Yurchenko, A., Goulas, J., Corre, R., Delacour, Q., Droin, N., Dessen, P., Goidin, D., Lange, S. S., Bhetawal, S., Mitjavila-Garcia, M. T., Baldacci, G., Nikolaev, S., Cadoret, J. C., Wood, R. D., & Kannouche, P. L. (2021). DNA polymerase zeta contributes to heterochromatin replication to prevent genome instability.
The EMBO Journal,
40(21), e104543.
https://doi.org/10.15252/embj.2020104543
Kychygina, A., Dall’Osto, M., Allen, J. A. M., Cadoret, J.-C., Piras, V., Pickett, H. A., & Crabbe, L. (2021). Progerin impairs 3D genome organization and induces fragile telomeres by limiting the dNTP pools.
Scientific Reports,
11(1), 13195.
https://doi.org/10.1038/s41598-021-92631-z
Marsin, S., Adam, Y., Cargemel, C., Andreani, J., Baconnais, S., Legrand, P., Li de la Sierra-Gallay, I., Humbert, A., Aumont-Nicaise, M., Velours, C., Ochsenbein, F., Durand, D., Le Cam, E., Walbott, H., Possoz, C., Quevillon-Cheruel, S., & Ferat, J.-L. (2021). Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase.
Nucleic Acids Research,
49(11), 6569–6586.
https://doi.org/10.1093/nar/gkab463
Courtot, L., Bournique, E., Maric, C., Guitton-Sert, L., Madrid-Mencía, M., Pancaldi, V., Cadoret, J.-C., Hoffmann, J.-S., & Bergoglio, V. (2021). Low Replicative Stress Triggers Cell-Type Specific Inheritable Advanced Replication Timing.
International Journal of Molecular Sciences,
22(9), 4959.
https://doi.org/10.3390/ijms22094959
Hadjadj, D., Denecker, T., Guérin, E., Kim, S.-J., Fauchereau, F., Baldacci, G., Maric, C., & Cadoret, J.-C. (2020). Efficient, quick and easy-to-use DNA replication timing analysis with START-R suite.
NAR Genomics and Bioinformatics,
2(2), lqaa045.
https://doi.org/10.1093/nargab/lqaa045
Hatem, E., Azzi, S., El Banna, N., He, T., Heneman-Masurel, A., Vernis, L., Baïlle, D., Masson, V., Dingli, F., Loew, D., Azzarone, B., Eid, P., Baldacci, G., & Huang, M.-E. (2019). Auranofin/Vitamin C: A Novel Drug Combination Targeting Triple-Negative Breast Cancer.
JNCI: Journal of the National Cancer Institute,
111(6), 597–608.
https://doi.org/10.1093/jnci/djy149
Gialluisi, A., Andlauer, T. F. M., Mirza-Schreiber, N., Moll, K., Becker, J., Hoffmann, P., Ludwig, K. U., Czamara, D., St Pourcain, B., Brandler, W., Honbolygó, F., Tóth, D., Csépe, V., Huguet, G., Morris, A. P., Hulslander, J., Willcutt, E. G., DeFries, J. C., Olson, R. K., … Schulte-Körne, G. (2019). Genome-wide association scan identifies new variants associated with a cognitive predictor of dyslexia.
Translational Psychiatry,
9(1), 1–15.
https://doi.org/10.1038/s41398-019-0402-0
Maes, P., Alkhovsky, S. V., Bào, Y., Beer, M., Birkhead, M., Briese, T., Buchmeier, M. J., Calisher, C. H., Charrel, R. N., Choi, I. R., Clegg, C. S., de la Torre, J. C., Delwart, E., DeRisi, J. L., Di Bello, P. L., Di Serio, F., Digiaro, M., Dolja, V. V., Drosten, C., … Kuhn, J. H. (2018). Taxonomy of the family Arenaviridae and the order Bunyavirales: update 2018.
Archives of Virology,
163(8), 2295–2310.
https://doi.org/10.1007/s00705-018-3843-5
Almeida, R., Fernández-Justel, J. M., Santa-María, C., Cadoret, J.-C., Cano-Aroca, L., Lombraña, R., Herranz, G., Agresti, A., & Gómez, M. (2018). Chromatin conformation regulates the coordination between DNA replication and transcription.
Nature Communications,
9(1), 1590.
https://doi.org/10.1038/s41467-018-03539-8
Hadjadj, D., Kim, S.-J., Denecker, T., Ben Driss, L., Cadoret, J.-C., Maric, C., Baldacci, G., & Fauchereau, F. (2017). A hypothesis-driven approach identifies CDK4 and CDK6 inhibitors as candidate drugs for treatments of adrenocortical carcinomas.
Aging,
9(12), 2695–2716.
https://doi.org/10.18632/aging.101356
Brustel, J., Kirstein, N., Izard, F., Grimaud, C., Prorok, P., Cayrou, C., Schotta, G., Abdelsamie, A. F., Déjardin, J., Méchali, M., Baldacci, G., Sardet, C., Cadoret, J.-C., Schepers, A., & Julien, E. (2017). Histone H4K20 tri-methylation at late-firing origins ensures timely heterochromatin replication.
The EMBO Journal,
36(18), 2726–2741.
https://doi.org/10.15252/embj.201796541
Pagan, C., Goubran-Botros, H., Delorme, R., Benabou, M., Lemière, N., Murray, K., Amsellem, F., Callebert, J., Chaste, P., Jamain, S., Fauchereau, F., Huguet, G., Maronde, E., Leboyer, M., Launay, J.-M., & Bourgeron, T. (2017). Disruption of melatonin synthesis is associated with impaired 14-3-3 and miR-451 levels in patients with autism spectrum disorders.
Scientific Reports,
7(1), 2096.
https://doi.org/10.1038/s41598-017-02152-x
Revue
2913254
WYW97AV7
1
apa
50
date
desc
8554
https://www.ijm.fr/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3Afalse%2C%22meta%22%3A%7B%22request_last%22%3A0%2C%22request_next%22%3A0%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%224LRC8TYG%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Briu%20et%20al.%22%2C%22parsedDate%22%3A%222021-01%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EBriu%2C%20L.-M.%2C%20Maric%2C%20C.%2C%20%26amp%3B%20Cadoret%2C%20J.-C.%20%282021%29.%20Replication%20Stress%2C%20Genomic%20Instability%2C%20and%20Replication%20Timing%3A%20A%20Complex%20Relationship.%20%3Ci%3EInternational%20Journal%20of%20Molecular%20Sciences%3C%5C%2Fi%3E%2C%20%3Ci%3E22%3C%5C%2Fi%3E%289%29%2C%204764.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fijms22094764%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fijms22094764%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Replication%20Stress%2C%20Genomic%20Instability%2C%20and%20Replication%20Timing%3A%20A%20Complex%20Relationship%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lina-Marie%22%2C%22lastName%22%3A%22Briu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Chrystelle%22%2C%22lastName%22%3A%22Maric%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Charles%22%2C%22lastName%22%3A%22Cadoret%22%7D%5D%2C%22abstractNote%22%3A%22The%20replication-timing%20program%20constitutes%20a%20key%20element%20of%20the%20organization%20and%20coordination%20of%20numerous%20nuclear%20processes%20in%20eukaryotes.%20This%20program%20is%20established%20at%20a%20crucial%20moment%20in%20the%20cell%20cycle%20and%20occurs%20simultaneously%20with%20the%20organization%20of%20the%20genome%2C%20thus%20indicating%20the%20vital%20significance%20of%20this%20process.%20With%20recent%20technological%20achievements%20of%20high-throughput%20approaches%2C%20a%20very%20strong%20link%20has%20been%20confirmed%20between%20replication%20timing%2C%20transcriptional%20activity%2C%20the%20epigenetic%20and%20mutational%20landscape%2C%20and%20the%203D%20organization%20of%20the%20genome.%20There%20is%20also%20a%20clear%20relationship%20between%20replication%20stress%2C%20replication%20timing%2C%20and%20genomic%20instability%2C%20but%20the%20extent%20to%20which%20they%20are%20mutually%20linked%20to%20each%20other%20is%20unclear.%20Recent%20evidence%20has%20shown%20that%20replication%20timing%20is%20affected%20in%20cancer%20cells%2C%20although%20the%20cause%20and%20consequence%20of%20this%20effect%20remain%20unknown.%20However%2C%20in-depth%20studies%20remain%20to%20be%20performed%20to%20characterize%20the%20molecular%20mechanisms%20of%20replication-timing%20regulation%20and%20clearly%20identify%20different%20cis-%20and%20trans-acting%20factors.%20The%20results%20of%20these%20studies%20will%20potentially%20facilitate%20the%20discovery%20of%20new%20therapeutic%20pathways%2C%20particularly%20for%20personalized%20medicine%2C%20or%20new%20biomarkers.%20This%20review%20focuses%20on%20the%20complex%20relationship%20between%20replication%20timing%2C%20replication%20stress%2C%20and%20genomic%20instability.%22%2C%22date%22%3A%222021%5C%2F1%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.3390%5C%2Fijms22094764%22%2C%22ISSN%22%3A%221422-0067%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.mdpi.com%5C%2F1422-0067%5C%2F22%5C%2F9%5C%2F4764%22%2C%22collections%22%3A%5B%22WYW97AV7%22%5D%2C%22dateModified%22%3A%222023-06-02T09%3A13%3A10Z%22%7D%7D%5D%7D
Briu, L.-M., Maric, C., & Cadoret, J.-C. (2021). Replication Stress, Genomic Instability, and Replication Timing: A Complex Relationship.
International Journal of Molecular Sciences,
22(9), 4764.
https://doi.org/10.3390/ijms22094764
Chapitre de livre
Manuel de Sciences fondamentales pour l’ostéopathe, chapitre sur la biologie moléculaire, Edition DE BOECK, parution août 2018, Auteur de deux chapitres : Biologie moléculaire et Biologie cellulaire
Logiciel
Logiciel START-R : https://github.com/thomasdenecker/START-R[/vc_column_text][/vc_tta_section][vc_tta_section title= »Thèses » tab_id= »1645024967580-08355b73-0edc »][vc_column_text]• Lina-Marie Briu (depuis 2020)
• Su-Jung Kim ( 2017-2021) → Post-doctorat à l’Institut Gustave Roussy
• Djihad Hadjad (2014-2018) → Poste de Maître de conférence à l’Université de Paris
 profil obtenu à partir d’un séquençage avec les anticorps anti pol epsilon et delta ainsi que celui obtenu sur l’échantillon Input
profil obtenu à partir d’un séquençage avec les anticorps anti pol epsilon et delta ainsi que celui obtenu sur l’échantillon Input




 Giuseppe BALDACCI, Enseignant-chercheur émérite, CADORET LAB+33 (0)1 57 27 80 73, bureau RB05B
Giuseppe BALDACCI, Enseignant-chercheur émérite, CADORET LAB+33 (0)1 57 27 80 73, bureau RB05B Samuel BENSOUSSAN, Stagiaire, CADORET LAB
Samuel BENSOUSSAN, Stagiaire, CADORET LAB Lina-Marie BRIU, Doctorante, CADORET LAB+33 (0)1 57 27 80 74, bureau 254B
Lina-Marie BRIU, Doctorante, CADORET LAB+33 (0)1 57 27 80 74, bureau 254B Chrystelle MARIC ANTOINAT, Chercheur, CADORET LAB+33 (0)1 57 27 80 73, bureau 254B
Chrystelle MARIC ANTOINAT, Chercheur, CADORET LAB+33 (0)1 57 27 80 73, bureau 254B