Recherche
Notre équipe de recherche développe et utilise des modèles in vitro reproduisant le développement normal et pathologique de la moelle épinière humaine et murine afin d’explorer les mécanismes fondamentaux qui le régissent. Nous concentrons nos efforts sur les modes d’action des voies de signalisation impliquant les FGFs, TGFβs et BMPs, ainsi que sur le rôle des facteurs de transcription HOX et PAX. Nos travaux visent également à mieux comprendre des pathologies associées : l’amyotrophie spinale infantile, le rhabdomyosarcome, le spina bifida et la sclérose latérale amyotrophique
Axe 1: PAX3/7 et BMPs & Régulation spatiale des destins et des morphologies cellulaires.
Les facteurs de transcription paralogues PAX3 et PAX7 peuvent agir à la fois comme activateurs et répresseurs transcriptionnels. Cette dualité fonctionnelle des PAX est modulée le long de l’axe dorso-ventral de la moelle épinière embryonnaire, par le gradient de signalisation par les BMPs, avec chacune des fonctionnalités déterminant des destins cellulaires et des morphologies spécifiques. Nous cherchons à décrypter comment cette activité bivalente est intégrée au niveau du génome, en analysant le recrutement des PAX et de leurs partenaires, ainsi que les dynamiques de la chromatine qui y sont associées. Parallèlement, nous étudions comment les réseaux de gènes régulés par les PAX orchestrent leurs effets sur le destin cellulaire, la morphologie et les propriétés mécaniques du neuroépithélium. Ces recherches visent à mieux comprendre comment des mutations touchant les PAX ou les gènes de leurs réseaux peuvent engendrer des défauts de fermeture du tube neural et de neurogenèse, à l’origine d’une pathologie congénitale majeure : le spina bifida.
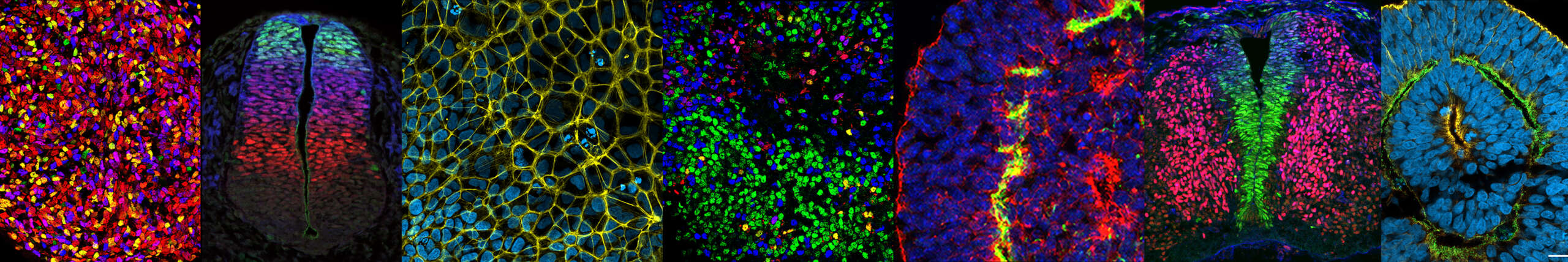
Axe 2: Dynamique de signalisation et spécification de la diversité neuronale.
Nous avons montré que les voies de signalisation des FGFs et de TGFβ rythment l’induction des gènes HOX au sein de leurs clusters génomiques dans les progéniteurs spinaux humains. Nous investiguons si ces mécanismes impliquent des dynamiques temporelles de l’activité des voies de signalisation. Par ailleurs, nous évaluons l’importance fonctionnelle de ces dynamiques dans la génération de la diversité cellulaire, qui sous-tend la formation des circuits locomoteurs. Pour ce faire, nous utilisons des modèles d’organoïdes innovants dans lesquels les gènes HOX sont induits de manière séquentielle dans des progéniteurs générant un tube neural. Celui-ci présente des profils de gènes HOX organisés en bandes le long de l’axe d’élongation, mimant l’organisation observée dans la moelle épinière embryonnaire.
Axe 3 : Bases cellulaires et moléculaires de maladie du motoneurons
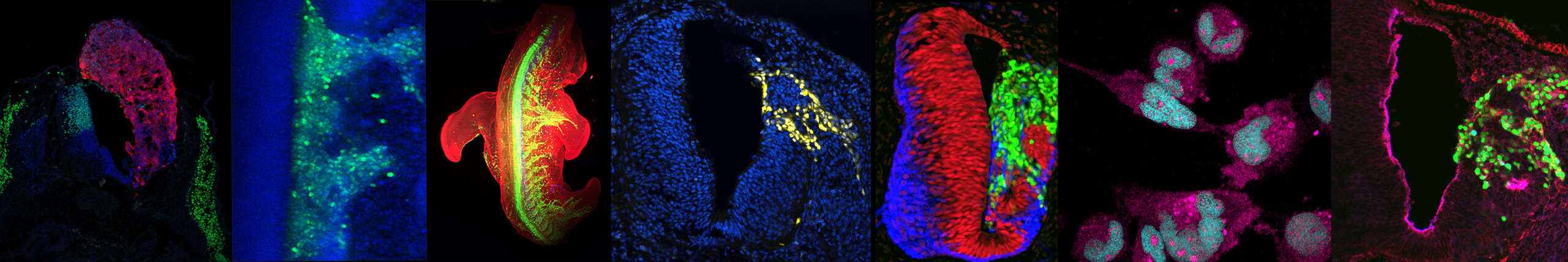
Ces modèles in vitro permettent d’accéder à des tissus ou des types cellulaires humains affectés dans des maladies. Nous utilisons donc des cellules souches pluripotentes induites dérivées de patients et ces modèles d’embryogenèse in vitro pour étudier les bases des maladies des motoneurones, un groupe hétérogène de maladies incurables et souvent mortelles. Nous étudions en particulier les atrophies musculaires spinales (SMAs) infantiles qui, bien que provoquées par des mutations dans des gènes exprimés de manière ubiquitaire, sont liées à des défauts de formation ou de survie de populations particulières de motoneurones tandis que d’autres sont préservées. Décrypter les bases de la vulnérabilité ou de la résistance de ces différents types de motoneurones pourrait ouvrir de nouvelles pistes thérapeutiques. Grâce à ces approches, nous commençons un projet sur la sclérose latérale amyotrophique en collaboration avec Odil Porrua.
Axe 4 : Détournement des signaux neurodéveloppementaux dans le rhabdomyosarcome
Le déploiement ectopique des voies de signalisation développementales dans les cancers pédiatriques, y compris le rhabdomyosarcome, pourrait expliquer leur évolution rapide sans accumulation des aberrations proto-oncogéniques typiques. Dans ce projet, nous évaluons le potentiel des cellules neuronales à donner naissance à des néoplasies rappelant le rhabdomyosarcome. En collaboration avec M. Castets (CRCL) et E. Pasquier (CRCM), nous étudions le rôle des signaux neurodéveloppementaux dans l’émergence de ce cancer et explorons la possibilité de cibler ces signaux pour induire une sensibilité des cellules de rhabdomyosarcome aux chimiothérapies déjà disponibles.
Membres
Responsables
Stephane NEDELEC,
Chercheur,
RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Vanessa RIBES,
Chercheur,
RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Membres
Kenza CHERIET,
Doctorante,
RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Claire DUGAST,
Chercheur,
RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Pascale GILARDI HEBENSTREIT,
Chercheur,
RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Marine GRISON,
Ingénieure en biologie,
RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Grace HENSTONE,
Doctorante,
RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Theaud HEZEZ,
Doctorant,
RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Helena MALEK,
Doctorante,
RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Camil MIRDASS,
Doctorant,
RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Robin RONDON,
Doctorant,
RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Pour contacter un membre de l’équipe par mail : prenom.nom@ijm.fr

Sélection de publication
Stem cell-derived models of spinal neurulation. Mirdass C, Catala M, Bocel M, Nedelec S, Ribes V. Emerg Top Life Sci. 2023 Dec 18;7(4):423-437. doi: 10.1042/ETLS20230087. PMID: 38087891 Review.
Self-organizing models of human trunk organogenesis recapitulate spinal cord and spine co-morphogenesis. Gribaudo S, Robert R, van Sambeek B, Mirdass C, Lyubimova A, Bouhali K, Ferent J, Morin X, van Oudenaarden A, Nedelec S. Nat Biotechnol. 2024 Aug;42(8):1243-1253. doi: 10.1038/s41587-023-01956-9. Epub 2023 Sep 14. PMID: 37709912
Single-cell transcriptomic analysis reveals diversity within mammalian spinal motor neurons. Liau ES, Jin S, Chen YC, Liu WS, Calon M, Nedelec S, Nie Q, Chen JA. Nat Commun. 2023 Jan 3;14(1):46. doi: 10.1038/s41467-022-35574-x. PMID: 36596814 Free PMC article.
Dynamic extrinsic pacing of the HOX clock in human axial progenitors controls motor neuron subtype specification. Mouilleau V, Vaslin C, Robert R, Gribaudo S, Nicolas N, Jarrige M, Terray A, Lesueur L, Mathis MW, Croft G, Daynac M, Rouiller-Fabre V, Wichterle H, Ribes V, Martinat C, Nedelec S. Development. 2021 Mar 29;148(6):dev194514. doi: 10.1242/dev.194514.
The PAX-FOXO1s trigger fast trans-differentiation of chick embryonic neural cells into alveolar rhabdomyosarcoma with tissue invasive properties limited by S phase entry inhibition. Gonzalez Curto G, Der Vartanian A, Frarma YE, Manceau L, Baldi L, Prisco S, Elarouci N, Causeret F, Korenkov D, Rigolet M, Aurade F, De Reynies A, Contremoulins V, Relaix F, Faklaris O, Briscoe J, Gilardi-Hebenstreit P, Ribes V. PLoS Genet. 2020 Nov 11;16(11):e1009164. doi: 10.1371/journal.pgen.1009164.
In vitro models of spinal motor circuit’s development in mammals: achievements and challenges. Nedelec S, Martinez-Arias A. Curr Opin Neurobiol. 2021 Feb;66:240-249. doi: 10.1016/j.conb.2020.12.002. Epub 2021 Mar 5. PMID: 33677159 Review.
Dullard-mediated Smad1/5/8 inhibition controls mouse cardiac neural crest cells condensation and outflow tract septation. Darrigrand JF, Valente M, Comai G, Martinez P, Petit M, Nishinakamura R, Osorio DS, Renault G, Marchiol C, Ribes V, Cadot B. Elife. 2020 Feb 27;9:e50325. doi: 10.7554/eLife.50325.
BMP4 patterns Smad activity and generates stereotyped cell fate organization in spinal organoids. Duval N, Vaslin C, Barata TC, Frarma Y, Contremoulins V, Baudin X, Nedelec S, Ribes VC. Development. 2019 Jul 25;146(14):dev175430. doi: 10.1242/dev.175430.
Pax3- and Pax7-mediated Dbx1 regulation orchestrates the patterning of intermediate spinal interneurons. Gard C, Gonzalez Curto G, Frarma YE, Chollet E, Duval N, Auzié V, Auradé F, Vigier L, Relaix F, Pierani A, Causeret F, Ribes V. Dev Biol. 2017 Dec 1;432(1):24-33. doi: 10.1016/j.ydbio.2017.06.014.
Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Maury Y, Côme J, Piskorowski RA, Salah-Mohellibi N, Chevaleyre V, Peschanski M, Martinat C, Nedelec S. Nat Biotechnol. 2015 Jan;33(1):89-96. doi: 10.1038/nbt.3049. Epub 2014 Nov 10. PMID: 25383599
Toutes les publications depuis 2017
Publications
2913254
MIYUW3RH
1
apa
50
date
desc
8845
https://www.ijm.fr/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3Afalse%2C%22meta%22%3A%7B%22request_last%22%3A0%2C%22request_next%22%3A0%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%22MZVS9E98%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Rondon%20et%20al.%22%2C%22parsedDate%22%3A%222025-03-12%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ERondon%2C%20R.%2C%20Hezez%2C%20T.%2C%20Albert%2C%20J.%20R.%2C%20Drayton-Libotte%2C%20B.%2C%20Gonzalez-Curto%2C%20G.%2C%20Relaix%2C%20F.%2C%20Dugast-Darzacq%2C%20C.%2C%20Gilardi-Hebenstreit%2C%20P.%2C%20%26amp%3B%20Ribes%2C%20V.%20%282025%29.%20%3Ci%3EDual%20PAX3%5C%2F7%20transcriptional%20activities%20spatially%20encode%20spinal%20cell%20fates%20through%20distinct%20gene%20networks%3C%5C%2Fi%3E.%20bioRxiv.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1101%5C%2F2025.03.11.642668%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1101%5C%2F2025.03.11.642668%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22preprint%22%2C%22title%22%3A%22Dual%20PAX3%5C%2F7%20transcriptional%20activities%20spatially%20encode%20spinal%20cell%20fates%20through%20distinct%20gene%20networks%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Robin%22%2C%22lastName%22%3A%22Rondon%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Th%5Cu00e9aud%22%2C%22lastName%22%3A%22Hezez%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julien%20Richard%22%2C%22lastName%22%3A%22Albert%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bernadette%22%2C%22lastName%22%3A%22Drayton-Libotte%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gloria%22%2C%22lastName%22%3A%22Gonzalez-Curto%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fr%5Cu00e9d%5Cu00e9ric%22%2C%22lastName%22%3A%22Relaix%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Claire%22%2C%22lastName%22%3A%22Dugast-Darzacq%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pascale%22%2C%22lastName%22%3A%22Gilardi-Hebenstreit%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Ribes%22%7D%5D%2C%22abstractNote%22%3A%22Understanding%20how%20transcription%20factors%20regulate%20organized%20cellular%20diversity%20in%20developing%20tissues%20remains%20a%20major%20challenge%20due%20to%20their%20pleiotropic%20functions.%20We%20addressed%20this%20by%20monitoring%20and%20genetically%20modulating%20the%20activity%20of%20PAX3%20and%20PAX7%20during%20the%20specification%20of%20neural%20progenitor%20pools%20in%20the%20embryonic%20spinal%20cord.%20Using%20mouse%20models%2C%20we%20show%20that%20the%20balance%20between%20the%20transcriptional%20activating%20and%20repressing%20functions%20of%20these%20factors%20is%20modulated%20along%20the%20dorsoventral%20axis%20and%20is%20instructive%20to%20the%20patterning%20of%20spinal%20progenitor%20pools.%20By%20combining%20loss-of-function%20experiments%20with%20functional%20genomics%20in%20spinal%20organoids%2C%20we%20demonstrate%20that%20PAX-mediated%20repression%20and%20activation%20rely%20on%20distinct%20cis-regulatory%20genomic%20modules.%20This%20enables%20both%20the%20coexistence%20of%20their%20dual%20activity%20in%20dorsal%20cell%20progenitors%20and%20the%20specific%20control%20of%20two%20major%20differentiation%20programs.%20PAX%20promotes%20H3K27me3%20deposition%20at%20silencers%20to%20repress%20ventral%20identities%2C%20while%20at%20enhancers%2C%20they%20act%20as%20pioneer%20factors%2C%20opening%20and%20activating%20cis-regulatory%20modules%20to%20specify%20dorsal-most%20identities.%20Finally%2C%20we%20show%20that%20this%20pioneer%20activity%20is%20restricted%20to%20cells%20exposed%20to%20BMP%20morphogens%2C%20ensuring%20spatial%20specificity.%20These%20findings%20reveal%20how%20PAX%20proteins%2C%20modulated%20by%20morphogen%20gradients%2C%20orchestrate%20neuronal%20diversity%20in%20the%20spinal%20cord%2C%20providing%20a%20robust%20framework%20for%20neural%20subtype%20specification.%22%2C%22genre%22%3A%22%22%2C%22repository%22%3A%22bioRxiv%22%2C%22archiveID%22%3A%22%22%2C%22date%22%3A%222025-03-12%22%2C%22DOI%22%3A%2210.1101%5C%2F2025.03.11.642668%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.biorxiv.org%5C%2Fcontent%5C%2F10.1101%5C%2F2025.03.11.642668v1%22%2C%22language%22%3A%22en%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-03-25T10%3A20%3A53Z%22%7D%7D%2C%7B%22key%22%3A%22V3P8MHUF%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Huchede%20et%20al.%22%2C%22parsedDate%22%3A%222024-10-07%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EHuchede%2C%20P.%2C%20Meyer%2C%20S.%2C%20Berthelot%2C%20C.%2C%20Hamadou%2C%20M.%2C%20Bertrand-Chapel%2C%20A.%2C%20Rakotomalala%2C%20A.%2C%20Manceau%2C%20L.%2C%20Tomine%2C%20J.%2C%20Lespinasse%2C%20N.%2C%20Lewandowski%2C%20P.%2C%20Cordier-Bussat%2C%20M.%2C%20Broutier%2C%20L.%2C%20Dutour%2C%20A.%2C%20Rochet%2C%20I.%2C%20Blay%2C%20J.-Y.%2C%20Degletagne%2C%20C.%2C%20Attignon%2C%20V.%2C%20Montero-Carcaboso%2C%20A.%2C%20Le%20Grand%2C%20M.%2C%20%26%23×2026%3B%20Castets%2C%20M.%20%282024%29.%20BMP2%20and%20BMP7%20cooperate%20with%20H3.3K27M%20to%20promote%20quiescence%20and%20invasiveness%20in%20pediatric%20diffuse%20midline%20gliomas.%20%3Ci%3EELife%3C%5C%2Fi%3E%2C%20%3Ci%3E12%3C%5C%2Fi%3E%2C%20RP91313.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.91313%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.91313%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22BMP2%20and%20BMP7%20cooperate%20with%20H3.3K27M%20to%20promote%20quiescence%20and%20invasiveness%20in%20pediatric%20diffuse%20midline%20gliomas%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Paul%22%2C%22lastName%22%3A%22Huchede%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Swann%22%2C%22lastName%22%3A%22Meyer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cl%5Cu00e9ment%22%2C%22lastName%22%3A%22Berthelot%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Maud%22%2C%22lastName%22%3A%22Hamadou%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Adrien%22%2C%22lastName%22%3A%22Bertrand-Chapel%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Andria%22%2C%22lastName%22%3A%22Rakotomalala%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Line%22%2C%22lastName%22%3A%22Manceau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julia%22%2C%22lastName%22%3A%22Tomine%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Lespinasse%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Paul%22%2C%22lastName%22%3A%22Lewandowski%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Martine%22%2C%22lastName%22%3A%22Cordier-Bussat%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laura%22%2C%22lastName%22%3A%22Broutier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Aur%5Cu00e9lie%22%2C%22lastName%22%3A%22Dutour%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Isabelle%22%2C%22lastName%22%3A%22Rochet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Yves%22%2C%22lastName%22%3A%22Blay%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cyril%22%2C%22lastName%22%3A%22Degletagne%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Val%5Cu00e9ry%22%2C%22lastName%22%3A%22Attignon%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Angel%22%2C%22lastName%22%3A%22Montero-Carcaboso%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marion%22%2C%22lastName%22%3A%22Le%20Grand%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eddy%22%2C%22lastName%22%3A%22Pasquier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alexandre%22%2C%22lastName%22%3A%22Vasiljevic%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pascale%22%2C%22lastName%22%3A%22Gilardi-Hebenstreit%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Samuel%22%2C%22lastName%22%3A%22Meignan%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pierre%22%2C%22lastName%22%3A%22Leblond%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Ribes%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Erika%22%2C%22lastName%22%3A%22Cosset%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marie%22%2C%22lastName%22%3A%22Castets%22%7D%5D%2C%22abstractNote%22%3A%22Pediatric%20diffuse%20midline%20gliomas%20%28pDMG%29%20are%20an%20aggressive%20type%20of%20childhood%20cancer%20with%20a%20fatal%20outcome.%20Their%20major%20epigenetic%20determinism%20has%20become%20clear%2C%20notably%20with%20the%20identification%20of%20K27M%20mutations%20in%20histone%20H3.%20However%2C%20the%20synergistic%20oncogenic%20mechanisms%20that%20induce%20and%20maintain%20tumor%20cell%20phenotype%20have%20yet%20to%20be%20deciphered.%20In%2020%20to%2030%25%20of%20cases%2C%20these%20tumors%20have%20an%20altered%20BMP%20signaling%20pathway%20with%20an%20oncogenic%20mutation%20on%20the%20BMP%20type%20I%20receptor%20ALK2%2C%20encoded%20by%20ACVR1.%20However%2C%20the%20potential%20impact%20of%20the%20BMP%20pathway%20in%20tumors%20non-mutated%20for%20ACVR1%20is%20less%20clear.%20By%20integrating%20bulk%2C%20single-cell%2C%20and%20spatial%20transcriptomic%20data%2C%20we%20show%20here%20that%20the%20BMP%20signaling%20pathway%20is%20activated%20at%20similar%20levels%20between%20ACVR1%20wild-type%20and%20mutant%20tumors%20and%20identify%20BMP2%20and%20BMP7%20as%20putative%20activators%20of%20the%20pathway%20in%20a%20specific%20subpopulation%20of%20cells.%20By%20using%20both%20pediatric%20isogenic%20glioma%20lines%20genetically%20modified%20to%20overexpress%20H3.3K27M%20and%20patients-derived%20DIPG%20cell%20lines%2C%20we%20demonstrate%20that%20BMP2%5C%2F7%20synergizes%20with%20H3.3K27M%20to%20induce%20a%20transcriptomic%20rewiring%20associated%20with%20a%20quiescent%20but%20invasive%20cell%20state.%20These%20data%20suggest%20a%20generic%20oncogenic%20role%20for%20the%20BMP%20pathway%20in%20gliomagenesis%20of%20pDMG%20and%20pave%20the%20way%20for%20specific%20targeting%20of%20downstream%20effectors%20mediating%20the%20K27M%5C%2FBMP%20crosstalk.%22%2C%22date%22%3A%222024-10-07%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.7554%5C%2FeLife.91313%22%2C%22ISSN%22%3A%222050-084X%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A48Z%22%7D%7D%2C%7B%22key%22%3A%22274JEH8E%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Rodr%5Cu00edguez-Mart%5Cu00edn%20et%20al.%22%2C%22parsedDate%22%3A%222024-04-10%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ERodr%26%23xED%3Bguez-Mart%26%23xED%3Bn%2C%20M.%2C%20B%26%23xE1%3Bez-Flores%2C%20J.%2C%20Ribes%2C%20V.%2C%20Isidoro-Garc%26%23xED%3Ba%2C%20M.%2C%20Lacal%2C%20J.%2C%20%26amp%3B%20Prieto-Matos%2C%20P.%20%282024%29.%20Non-Mammalian%20Models%20for%20Understanding%20Neurological%20Defects%20in%20RASopathies.%20%3Ci%3EBiomedicines%3C%5C%2Fi%3E%2C%20%3Ci%3E12%3C%5C%2Fi%3E%284%29%2C%20841.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fbiomedicines12040841%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fbiomedicines12040841%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Non-Mammalian%20Models%20for%20Understanding%20Neurological%20Defects%20in%20RASopathies%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mario%22%2C%22lastName%22%3A%22Rodr%5Cu00edguez-Mart%5Cu00edn%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Juan%22%2C%22lastName%22%3A%22B%5Cu00e1ez-Flores%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Ribes%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mar%5Cu00eda%22%2C%22lastName%22%3A%22Isidoro-Garc%5Cu00eda%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jesus%22%2C%22lastName%22%3A%22Lacal%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pablo%22%2C%22lastName%22%3A%22Prieto-Matos%22%7D%5D%2C%22abstractNote%22%3A%22RASopathies%2C%20a%20group%20of%20neurodevelopmental%20congenital%20disorders%20stemming%20from%20mutations%20in%20the%20RAS%5C%2FMAPK%20pathway%2C%20present%20a%20unique%20opportunity%20to%20delve%20into%20the%20intricacies%20of%20complex%20neurological%20disorders.%20Afflicting%20approximately%20one%20in%20a%20thousand%20newborns%2C%20RASopathies%20manifest%20as%20abnormalities%20across%20multiple%20organ%20systems%2C%20with%20a%20pronounced%20impact%20on%20the%20central%20and%20peripheral%20nervous%20system.%20In%20the%20pursuit%20of%20understanding%20RASopathies%27%20neurobiology%20and%20establishing%20phenotype-genotype%20relationships%2C%20in%20vivo%20non-mammalian%20models%20have%20emerged%20as%20indispensable%20tools.%20Species%20such%20as%20Danio%20rerio%2C%20Drosophila%20melanogaster%2C%20Caenorhabditis%20elegans%2C%20Xenopus%20species%20and%20Gallus%20gallus%20embryos%20have%20proven%20to%20be%20invaluable%20in%20shedding%20light%20on%20the%20intricate%20pathways%20implicated%20in%20RASopathies.%20Despite%20some%20inherent%20weaknesses%2C%20these%20genetic%20models%20offer%20distinct%20advantages%20over%20traditional%20rodent%20models%2C%20providing%20a%20holistic%20perspective%20on%20complex%20genetics%2C%20multi-organ%20involvement%2C%20and%20the%20interplay%20among%20various%20pathway%20components%2C%20offering%20insights%20into%20the%20pathophysiological%20aspects%20of%20mutations-driven%20symptoms.%20This%20review%20underscores%20the%20value%20of%20investigating%20the%20genetic%20basis%20of%20RASopathies%20for%20unraveling%20the%20underlying%20mechanisms%20contributing%20to%20broader%20neurological%20complexities.%20It%20also%20emphasizes%20the%20pivotal%20role%20of%20non-mammalian%20models%20in%20serving%20as%20a%20crucial%20preliminary%20step%20for%20the%20development%20of%20innovative%20therapeutic%20strategies.%22%2C%22date%22%3A%222024-04-10%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.3390%5C%2Fbiomedicines12040841%22%2C%22ISSN%22%3A%222227-9059%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A48Z%22%7D%7D%2C%7B%22key%22%3A%22SNLMHYWE%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Mirdass%20et%20al.%22%2C%22parsedDate%22%3A%222023-12-18%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EMirdass%2C%20C.%2C%20Catala%2C%20M.%2C%20Bocel%2C%20M.%2C%20Nedelec%2C%20S.%2C%20%26amp%3B%20Ribes%2C%20V.%20%282023%29.%20Stem%20cell-derived%20models%20of%20spinal%20neurulation.%20%3Ci%3EEmerging%20Topics%20in%20Life%20Sciences%3C%5C%2Fi%3E%2C%20%3Ci%3E7%3C%5C%2Fi%3E%284%29%2C%20423%26%23×2013%3B437.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1042%5C%2FETLS20230087%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1042%5C%2FETLS20230087%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Stem%20cell-derived%20models%20of%20spinal%20neurulation%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Camil%22%2C%22lastName%22%3A%22Mirdass%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Martin%22%2C%22lastName%22%3A%22Catala%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mika%5Cu00eblle%22%2C%22lastName%22%3A%22Bocel%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phane%22%2C%22lastName%22%3A%22Nedelec%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Ribes%22%7D%5D%2C%22abstractNote%22%3A%22Neurulation%20is%20a%20critical%20step%20in%20early%20embryonic%20development%2C%20giving%20rise%20to%20the%20neural%20tube%2C%20the%20primordium%20of%20the%20central%20nervous%20system%20in%20amniotes.%20Understanding%20this%20complex%2C%20multi-scale%2C%20multi-tissue%20morphogenetic%20process%20is%20essential%20to%20provide%20insights%20into%20normal%20development%20and%20the%20etiology%20of%20neural%20tube%20defects.%20Innovations%20in%20tissue%20engineering%20have%20fostered%20the%20generation%20of%20pluripotent%20stem%20cell-based%20in%20vitro%20models%2C%20including%20organoids%2C%20that%20are%20emerging%20as%20unique%20tools%20for%20delving%20into%20neurulation%20mechanisms%2C%20especially%20in%20the%20context%20of%20human%20development.%20Each%20model%20captures%20specific%20aspects%20of%20neural%20tube%20morphogenesis%2C%20from%20epithelialization%20to%20neural%20tissue%20elongation%2C%20folding%20and%20cavitation.%20In%20particular%2C%20the%20recent%20models%20of%20human%20and%20mouse%20trunk%20morphogenesis%2C%20such%20as%20gastruloids%2C%20that%20form%20a%20spinal%20neural%20plate-like%20or%20neural%20tube-like%20structure%20are%20opening%20new%20avenues%20to%20study%20normal%20and%20pathological%20neurulation.%20Here%2C%20we%20review%20the%20morphogenetic%20events%20generating%20the%20neural%20tube%20in%20the%20mammalian%20embryo%20and%20questions%20that%20remain%20unanswered.%20We%20discuss%20the%20advantages%20and%20limitations%20of%20existing%20in%20vitro%20models%20of%20neurulation%20and%20possible%20future%20technical%20developments.%22%2C%22date%22%3A%222023-12-18%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1042%5C%2FETLS20230087%22%2C%22ISSN%22%3A%222397-8554%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A48Z%22%7D%7D%2C%7B%22key%22%3A%22PAS5QN45%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Gribaudo%20et%20al.%22%2C%22parsedDate%22%3A%222023-09-14%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EGribaudo%2C%20S.%2C%20Robert%2C%20R.%2C%20van%20Sambeek%2C%20B.%2C%20Mirdass%2C%20C.%2C%20Lyubimova%2C%20A.%2C%20Bouhali%2C%20K.%2C%20Ferent%2C%20J.%2C%20Morin%2C%20X.%2C%20van%20Oudenaarden%2C%20A.%2C%20%26amp%3B%20Nedelec%2C%20S.%20%282023%29.%20Self-organizing%20models%20of%20human%20trunk%20organogenesis%20recapitulate%20spinal%20cord%20and%20spine%20co-morphogenesis.%20%3Ci%3ENature%20Biotechnology%3C%5C%2Fi%3E.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41587-023-01956-9%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41587-023-01956-9%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Self-organizing%20models%20of%20human%20trunk%20organogenesis%20recapitulate%20spinal%20cord%20and%20spine%20co-morphogenesis%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Simona%22%2C%22lastName%22%3A%22Gribaudo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R%5Cu00e9mi%22%2C%22lastName%22%3A%22Robert%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bj%5Cu00f6rn%22%2C%22lastName%22%3A%22van%20Sambeek%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Camil%22%2C%22lastName%22%3A%22Mirdass%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anna%22%2C%22lastName%22%3A%22Lyubimova%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Kamal%22%2C%22lastName%22%3A%22Bouhali%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julien%22%2C%22lastName%22%3A%22Ferent%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Xavier%22%2C%22lastName%22%3A%22Morin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alexander%22%2C%22lastName%22%3A%22van%20Oudenaarden%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phane%22%2C%22lastName%22%3A%22Nedelec%22%7D%5D%2C%22abstractNote%22%3A%22Integrated%20in%20vitro%20models%20of%20human%20organogenesis%20are%20needed%20to%20elucidate%20the%20multi-systemic%20events%20underlying%20development%20and%20disease.%20Here%20we%20report%20the%20generation%20of%20human%20trunk-like%20structures%20that%20model%20the%20co-morphogenesis%2C%20patterning%20and%20differentiation%20of%20the%20human%20spine%20and%20spinal%20cord.%20We%20identified%20differentiation%20conditions%20for%20human%20pluripotent%20stem%20cells%20favoring%20the%20formation%20of%20an%20embryo-like%20extending%20antero-posterior%20%28AP%29%20axis.%20Single-cell%20and%20spatial%20transcriptomics%20show%20that%20somitic%20and%20spinal%20cord%20differentiation%20trajectories%20organize%20along%20this%20axis%20and%20can%20self-assemble%20into%20a%20neural%20tube%20surrounded%20by%20somites%20upon%20extracellular%20matrix%20addition.%20Morphogenesis%20is%20coupled%20with%20AP%20patterning%20mechanisms%2C%20which%20results%2C%20at%20later%20stages%20of%20organogenesis%2C%20in%20in%20vivo-like%20arrays%20of%20neural%20subtypes%20along%20a%20neural%20tube%20surrounded%20by%20spine%20and%20muscle%20progenitors%20contacted%20by%20neuronal%20projections.%20This%20integrated%20system%20of%20trunk%20development%20indicates%20that%20in%20vivo-like%20multi-tissue%20co-morphogenesis%20and%20topographic%20organization%20of%20terminal%20cell%20types%20can%20be%20achieved%20in%20human%20organoids%2C%20opening%20windows%20for%20the%20development%20of%20more%20complex%20models%20of%20organogenesis.%22%2C%22date%22%3A%222023-09-14%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41587-023-01956-9%22%2C%22ISSN%22%3A%221546-1696%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A48Z%22%7D%7D%2C%7B%22key%22%3A%22RZ5P3XKY%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Liau%20et%20al.%22%2C%22parsedDate%22%3A%222023-01-03%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ELiau%2C%20E.%20S.%2C%20Jin%2C%20S.%2C%20Chen%2C%20Y.-C.%2C%20Liu%2C%20W.-S.%2C%20Calon%2C%20M.%2C%20Nedelec%2C%20S.%2C%20Nie%2C%20Q.%2C%20%26amp%3B%20Chen%2C%20J.-A.%20%282023%29.%20Single-cell%20transcriptomic%20analysis%20reveals%20diversity%20within%20mammalian%20spinal%20motor%20neurons.%20%3Ci%3ENature%20Communications%3C%5C%2Fi%3E%2C%20%3Ci%3E14%3C%5C%2Fi%3E%281%29%2C%2046.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41467-022-35574-x%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41467-022-35574-x%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Single-cell%20transcriptomic%20analysis%20reveals%20diversity%20within%20mammalian%20spinal%20motor%20neurons%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ee%20Shan%22%2C%22lastName%22%3A%22Liau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Suoqin%22%2C%22lastName%22%3A%22Jin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yen-Chung%22%2C%22lastName%22%3A%22Chen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Wei-Szu%22%2C%22lastName%22%3A%22Liu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ma%5Cu00ebliss%22%2C%22lastName%22%3A%22Calon%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phane%22%2C%22lastName%22%3A%22Nedelec%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Qing%22%2C%22lastName%22%3A%22Nie%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jun-An%22%2C%22lastName%22%3A%22Chen%22%7D%5D%2C%22abstractNote%22%3A%22Spinal%20motor%20neurons%20%28MNs%29%20integrate%20sensory%20stimuli%20and%20brain%20commands%20to%20generate%20movements.%20In%20vertebrates%2C%20the%20molecular%20identities%20of%20the%20cardinal%20MN%20types%20such%20as%20those%20innervating%20limb%20versus%20trunk%20muscles%20are%20well%20elucidated.%20Yet%20the%20identities%20of%20finer%20subtypes%20within%20these%20cell%20populations%20that%20innervate%20individual%20muscle%20groups%20remain%20enigmatic.%20Here%20we%20investigate%20heterogeneity%20in%20mouse%20MNs%20using%20single-cell%20transcriptomics.%20Among%20limb-innervating%20MNs%2C%20we%20reveal%20a%20diverse%20neuropeptide%20code%20for%20delineating%20putative%20motor%20pool%20identities.%20Additionally%2C%20we%20uncover%20that%20axial%20MNs%20are%20subdivided%20into%20three%20molecularly%20distinct%20subtypes%2C%20defined%20by%20mediolaterally-biased%20Satb2%2C%20Nr2f2%20or%20Bcl11b%20expression%20patterns%20with%20different%20axon%20guidance%20signatures.%20These%20three%20subtypes%20are%20present%20in%20chicken%20and%20human%20embryos%2C%20suggesting%20a%20conserved%20axial%20MN%20expression%20pattern%20across%20higher%20vertebrates.%20Overall%2C%20our%20study%20provides%20a%20molecular%20resource%20of%20spinal%20MN%20types%20and%20paves%20the%20way%20towards%20deciphering%20how%20neuronal%20subtypes%20evolved%20to%20accommodate%20vertebrate%20motor%20behaviors.%22%2C%22date%22%3A%222023-01-03%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41467-022-35574-x%22%2C%22ISSN%22%3A%222041-1723%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A48Z%22%7D%7D%2C%7B%22key%22%3A%22XKLFGQ38%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Manceau%20et%20al.%22%2C%22parsedDate%22%3A%222022-05%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EManceau%2C%20L.%2C%20Richard%20Albert%2C%20J.%2C%20Lollini%2C%20P.-L.%2C%20Greenberg%2C%20M.%20V.%20C.%2C%20Gilardi-Hebenstreit%2C%20P.%2C%20%26amp%3B%20Ribes%2C%20V.%20%282022%29.%20Divergent%20transcriptional%20and%20transforming%20properties%20of%20PAX3-FOXO1%20and%20PAX7-FOXO1%20paralogs.%20%3Ci%3EPLoS%20Genetics%3C%5C%2Fi%3E%2C%20%3Ci%3E18%3C%5C%2Fi%3E%285%29%2C%20e1009782.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1371%5C%2Fjournal.pgen.1009782%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1371%5C%2Fjournal.pgen.1009782%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Divergent%20transcriptional%20and%20transforming%20properties%20of%20PAX3-FOXO1%20and%20PAX7-FOXO1%20paralogs%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Line%22%2C%22lastName%22%3A%22Manceau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julien%22%2C%22lastName%22%3A%22Richard%20Albert%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pier-Luigi%22%2C%22lastName%22%3A%22Lollini%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Maxim%20V.%20C.%22%2C%22lastName%22%3A%22Greenberg%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pascale%22%2C%22lastName%22%3A%22Gilardi-Hebenstreit%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Ribes%22%7D%5D%2C%22abstractNote%22%3A%22The%20hallmarks%20of%20the%20alveolar%20subclass%20of%20rhabdomyosarcoma%20are%20chromosomal%20translocations%20that%20generate%20chimeric%20PAX3-FOXO1%20or%20PAX7-FOXO1%20transcription%20factors.%20Overexpression%20of%20either%20PAX-FOXO1s%20results%20in%20related%20cell%20transformation%20in%20animal%20models.%20Yet%2C%20in%20patients%20the%20two%20structural%20genetic%20aberrations%20they%20derived%20from%20are%20associated%20with%20distinct%20pathological%20manifestations.%20To%20assess%20the%20mechanisms%20underlying%20these%20differences%2C%20we%20generated%20isogenic%20fibroblast%20lines%20expressing%20either%20PAX-FOXO1%20paralog.%20Mapping%20of%20their%20genomic%20recruitment%20using%20CUT%26Tag%20revealed%20that%20the%20two%20chimeric%20proteins%20have%20distinct%20DNA%20binding%20preferences.%20In%20addition%2C%20PAX7-FOXO1%20binding%20results%20in%20greater%20recruitment%20of%20the%20H3K27ac%20activation%20mark%20than%20PAX3-FOXO1%20binding%20and%20is%20accompanied%20by%20greater%20transcriptional%20activation%20of%20neighbouring%20genes.%20These%20effects%20are%20associated%20with%20a%20PAX-FOXO1-specific%20alteration%20in%20the%20expression%20of%20genes%20regulating%20cell%20shape%20and%20the%20cell%20cycle.%20Consistently%2C%20PAX3-FOXO1%20accentuates%20fibroblast%20cellular%20traits%20associated%20with%20contractility%20and%20surface%20adhesion%20and%20limits%20entry%20into%20S%20phase.%20In%20contrast%2C%20PAX7-FOXO1%20drives%20cells%20to%20adopt%20an%20amoeboid%20shape%2C%20reduces%20entry%20into%20M%20phase%2C%20and%20causes%20increased%20DNA%20damage.%20Altogether%2C%20our%20results%20argue%20that%20the%20diversity%20of%20rhabdomyosarcoma%20manifestation%20arises%2C%20in%20part%2C%20from%20the%20divergence%20between%20the%20genomic%20occupancy%20and%20transcriptional%20activity%20of%20PAX3-FOXO1%20and%20PAX7-FOXO1.%22%2C%22date%22%3A%222022-05%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1371%5C%2Fjournal.pgen.1009782%22%2C%22ISSN%22%3A%221553-7404%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A48Z%22%7D%7D%2C%7B%22key%22%3A%229HV5ZNAP%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Mouilleau%20et%20al.%22%2C%22parsedDate%22%3A%222021-03-29%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EMouilleau%2C%20V.%2C%20Vaslin%2C%20C.%2C%20Robert%2C%20R.%2C%20Gribaudo%2C%20S.%2C%20Nicolas%2C%20N.%2C%20Jarrige%2C%20M.%2C%20Terray%2C%20A.%2C%20Lesueur%2C%20L.%2C%20Mathis%2C%20M.%20W.%2C%20Croft%2C%20G.%2C%20Daynac%2C%20M.%2C%20Rouiller-Fabre%2C%20V.%2C%20Wichterle%2C%20H.%2C%20Ribes%2C%20V.%2C%20Martinat%2C%20C.%2C%20%26amp%3B%20Nedelec%2C%20S.%20%282021%29.%20Dynamic%20extrinsic%20pacing%20of%20the%20HOX%20clock%20in%20human%20axial%20progenitors%20controls%20motor%20neuron%20subtype%20specification.%20%3Ci%3EDevelopment%20%28Cambridge%2C%20England%29%3C%5C%2Fi%3E%2C%20%3Ci%3E148%3C%5C%2Fi%3E%286%29%2C%20dev194514.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1242%5C%2Fdev.194514%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1242%5C%2Fdev.194514%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Dynamic%20extrinsic%20pacing%20of%20the%20HOX%20clock%20in%20human%20axial%20progenitors%20controls%20motor%20neuron%20subtype%20specification%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vincent%22%2C%22lastName%22%3A%22Mouilleau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C%5Cu00e9lia%22%2C%22lastName%22%3A%22Vaslin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R%5Cu00e9mi%22%2C%22lastName%22%3A%22Robert%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Simona%22%2C%22lastName%22%3A%22Gribaudo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nour%22%2C%22lastName%22%3A%22Nicolas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Margot%22%2C%22lastName%22%3A%22Jarrige%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ang%5Cu00e9lique%22%2C%22lastName%22%3A%22Terray%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L%5Cu00e9a%22%2C%22lastName%22%3A%22Lesueur%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mackenzie%20W.%22%2C%22lastName%22%3A%22Mathis%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gist%22%2C%22lastName%22%3A%22Croft%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mathieu%22%2C%22lastName%22%3A%22Daynac%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Virginie%22%2C%22lastName%22%3A%22Rouiller-Fabre%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hynek%22%2C%22lastName%22%3A%22Wichterle%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Ribes%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C%5Cu00e9cile%22%2C%22lastName%22%3A%22Martinat%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phane%22%2C%22lastName%22%3A%22Nedelec%22%7D%5D%2C%22abstractNote%22%3A%22Rostro-caudal%20patterning%20of%20vertebrates%20depends%20on%20the%20temporally%20progressive%20activation%20of%20HOX%20genes%20within%20axial%20stem%20cells%20that%20fuel%20axial%20embryo%20elongation.%20Whether%20the%20pace%20of%20sequential%20activation%20of%20HOX%20genes%2C%20the%20%27HOX%20clock%27%2C%20is%20controlled%20by%20intrinsic%20chromatin-based%20timing%20mechanisms%20or%20by%20temporal%20changes%20in%20extrinsic%20cues%20remains%20unclear.%20Here%2C%20we%20studied%20HOX%20clock%20pacing%20in%20human%20pluripotent%20stem%20cell-derived%20axial%20progenitors%20differentiating%20into%20diverse%20spinal%20cord%20motor%20neuron%20subtypes.%20We%20show%20that%20the%20progressive%20activation%20of%20caudal%20HOX%20genes%20is%20controlled%20by%20a%20dynamic%20increase%20in%20FGF%20signaling.%20Blocking%20the%20FGF%20pathway%20stalled%20induction%20of%20HOX%20genes%2C%20while%20a%20precocious%20increase%20of%20FGF%2C%20alone%20or%20with%20GDF11%20ligand%2C%20accelerated%20the%20HOX%20clock.%20Cells%20differentiated%20under%20accelerated%20HOX%20induction%20generated%20appropriate%20posterior%20motor%20neuron%20subtypes%20found%20along%20the%20human%20embryonic%20spinal%20cord.%20The%20pacing%20of%20the%20HOX%20clock%20is%20thus%20dynamically%20regulated%20by%20exposure%20to%20secreted%20cues.%20Its%20manipulation%20by%20extrinsic%20factors%20provides%20synchronized%20access%20to%20multiple%20human%20neuronal%20subtypes%20of%20distinct%20rostro-caudal%20identities%20for%20basic%20and%20translational%20applications.This%20article%20has%20an%20associated%20%27The%20people%20behind%20the%20papers%27%20interview.%22%2C%22date%22%3A%222021-03-29%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1242%5C%2Fdev.194514%22%2C%22ISSN%22%3A%221477-9129%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A47Z%22%7D%7D%2C%7B%22key%22%3A%22A7IZNFXP%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Nedelec%20and%20Martinez-Arias%22%2C%22parsedDate%22%3A%222021-02%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ENedelec%2C%20S.%2C%20%26amp%3B%20Martinez-Arias%2C%20A.%20%282021%29.%20In%20vitro%20models%20of%20spinal%20motor%20circuit%26%23×2019%3Bs%20development%20in%20mammals%3A%20achievements%20and%20challenges.%20%3Ci%3ECurrent%20Opinion%20in%20Neurobiology%3C%5C%2Fi%3E%2C%20%3Ci%3E66%3C%5C%2Fi%3E%2C%20240%26%23×2013%3B249.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.conb.2020.12.002%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.conb.2020.12.002%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22In%20vitro%20models%20of%20spinal%20motor%20circuit%27s%20development%20in%20mammals%3A%20achievements%20and%20challenges%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phane%22%2C%22lastName%22%3A%22Nedelec%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alfonso%22%2C%22lastName%22%3A%22Martinez-Arias%22%7D%5D%2C%22abstractNote%22%3A%22The%20connectivity%20patterns%20of%20neurons%20sustaining%20the%20functionality%20of%20spinal%20locomotor%20circuits%20rely%20on%20the%20specification%20of%20hundreds%20of%20motor%20neuron%20and%20interneuron%20subtypes%20precisely%20arrayed%20within%20the%20embryonic%20spinal%20cord.%20Knowledge%20acquired%20by%20developmental%20biologists%20on%20the%20molecular%20mechanisms%20underpinning%20this%20process%20in%20vivo%20has%20supported%20the%20development%20of%202D%20and%203D%20differentiation%20strategies%20to%20generate%20spinal%20neuronal%20diversity%20from%20mouse%20and%20human%20pluripotent%20stem%20cells%20%28PSCs%29.%20Here%2C%20we%20review%20recent%20breakthroughs%20in%20this%20field%20and%20the%20perspectives%20opened%20up%20by%20models%20of%20in%20vitro%20embryogenesis%20to%20approach%20the%20mechanisms%20underlying%20neuronal%20diversification%20and%20the%20formation%20of%20functional%20mouse%20and%20human%20locomotor%20circuits.%20Beyond%20serving%20fundamental%20investigations%2C%20these%20new%20approaches%20should%20help%20engineering%20neuronal%20circuits%20differentially%20impacted%20in%20neuromuscular%20disorders%2C%20such%20as%20amyotrophic%20lateral%20sclerosis%20or%20spinal%20muscular%20atrophies%2C%20and%20thus%20open%20new%20avenues%20for%20disease%20modeling%20and%20drug%20screenings.%22%2C%22date%22%3A%222021-02%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.conb.2020.12.002%22%2C%22ISSN%22%3A%221873-6882%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A47Z%22%7D%7D%2C%7B%22key%22%3A%22URU7A59T%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Gonzalez%20Curto%20et%20al.%22%2C%22parsedDate%22%3A%222020-11%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EGonzalez%20Curto%2C%20G.%2C%20Der%20Vartanian%2C%20A.%2C%20Frarma%2C%20Y.%20E.-M.%2C%20Manceau%2C%20L.%2C%20Baldi%2C%20L.%2C%20Prisco%2C%20S.%2C%20Elarouci%2C%20N.%2C%20Causeret%2C%20F.%2C%20Korenkov%2C%20D.%2C%20Rigolet%2C%20M.%2C%20Aurade%2C%20F.%2C%20De%20Reynies%2C%20A.%2C%20Contremoulins%2C%20V.%2C%20Relaix%2C%20F.%2C%20Faklaris%2C%20O.%2C%20Briscoe%2C%20J.%2C%20Gilardi-Hebenstreit%2C%20P.%2C%20%26amp%3B%20Ribes%2C%20V.%20%282020%29.%20The%20PAX-FOXO1s%20trigger%20fast%20trans-differentiation%20of%20chick%20embryonic%20neural%20cells%20into%20alveolar%20rhabdomyosarcoma%20with%20tissue%20invasive%20properties%20limited%20by%20S%20phase%20entry%20inhibition.%20%3Ci%3EPLoS%20Genetics%3C%5C%2Fi%3E%2C%20%3Ci%3E16%3C%5C%2Fi%3E%2811%29%2C%20e1009164.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1371%5C%2Fjournal.pgen.1009164%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1371%5C%2Fjournal.pgen.1009164%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20PAX-FOXO1s%20trigger%20fast%20trans-differentiation%20of%20chick%20embryonic%20neural%20cells%20into%20alveolar%20rhabdomyosarcoma%20with%20tissue%20invasive%20properties%20limited%20by%20S%20phase%20entry%20inhibition%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gloria%22%2C%22lastName%22%3A%22Gonzalez%20Curto%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Audrey%22%2C%22lastName%22%3A%22Der%20Vartanian%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Youcef%20El-Mokhtar%22%2C%22lastName%22%3A%22Frarma%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Line%22%2C%22lastName%22%3A%22Manceau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lorenzo%22%2C%22lastName%22%3A%22Baldi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Selene%22%2C%22lastName%22%3A%22Prisco%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nabila%22%2C%22lastName%22%3A%22Elarouci%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fr%5Cu00e9d%5Cu00e9ric%22%2C%22lastName%22%3A%22Causeret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Daniil%22%2C%22lastName%22%3A%22Korenkov%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Muriel%22%2C%22lastName%22%3A%22Rigolet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fr%5Cu00e9d%5Cu00e9ric%22%2C%22lastName%22%3A%22Aurade%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Aur%5Cu00e9lien%22%2C%22lastName%22%3A%22De%20Reynies%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vincent%22%2C%22lastName%22%3A%22Contremoulins%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fr%5Cu00e9d%5Cu00e9ric%22%2C%22lastName%22%3A%22Relaix%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Orestis%22%2C%22lastName%22%3A%22Faklaris%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22James%22%2C%22lastName%22%3A%22Briscoe%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pascale%22%2C%22lastName%22%3A%22Gilardi-Hebenstreit%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Ribes%22%7D%5D%2C%22abstractNote%22%3A%22The%20chromosome%20translocations%20generating%20PAX3-FOXO1%20and%20PAX7-FOXO1%20chimeric%20proteins%20are%20the%20primary%20hallmarks%20of%20the%20paediatric%20fusion-positive%20alveolar%20subtype%20of%20Rhabdomyosarcoma%20%28FP-RMS%29.%20Despite%20the%20ability%20of%20these%20transcription%20factors%20to%20remodel%20chromatin%20landscapes%20and%20promote%20the%20expression%20of%20tumour%20driver%20genes%2C%20they%20only%20inefficiently%20promote%20malignant%20transformation%20in%20vivo.%20The%20reason%20for%20this%20is%20unclear.%20To%20address%20this%2C%20we%20developed%20an%20in%20ovo%20model%20to%20follow%20the%20response%20of%20spinal%20cord%20progenitors%20to%20PAX-FOXO1s.%20Our%20data%20demonstrate%20that%20PAX-FOXO1s%2C%20but%20not%20wild-type%20PAX3%20or%20PAX7%2C%20trigger%20the%20trans-differentiation%20of%20neural%20cells%20into%20FP-RMS-like%20cells%20with%20myogenic%20characteristics.%20In%20parallel%2C%20PAX-FOXO1s%20remodel%20the%20neural%20pseudo-stratified%20epithelium%20into%20a%20cohesive%20mesenchyme%20capable%20of%20tissue%20invasion.%20Surprisingly%2C%20expression%20of%20PAX-FOXO1s%2C%20similar%20to%20wild-type%20PAX3%5C%2F7%2C%20reduce%20the%20levels%20of%20CDK-CYCLIN%20activity%20and%20increase%20the%20fraction%20of%20cells%20in%20G1.%20Introduction%20of%20CYCLIN%20D1%20or%20MYCN%20overcomes%20this%20PAX-FOXO1-mediated%20cell%20cycle%20inhibition%20and%20promotes%20tumour%20growth.%20Together%2C%20our%20findings%20reveal%20a%20mechanism%20that%20can%20explain%20the%20apparent%20limited%20oncogenicity%20of%20PAX-FOXO1%20fusion%20transcription%20factors.%20They%20are%20also%20consistent%20with%20certain%20clinical%20reports%20indicative%20of%20a%20neural%20origin%20of%20FP-RMS.%22%2C%22date%22%3A%222020-11%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1371%5C%2Fjournal.pgen.1009164%22%2C%22ISSN%22%3A%221553-7404%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A48Z%22%7D%7D%2C%7B%22key%22%3A%222VKRM6UL%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kumamoto%20et%20al.%22%2C%22parsedDate%22%3A%222020-08-19%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKumamoto%2C%20T.%2C%20Maurinot%2C%20F.%2C%20Barry-Martinet%2C%20R.%2C%20Vaslin%2C%20C.%2C%20Vandormael-Pournin%2C%20S.%2C%20Le%2C%20M.%2C%20Lerat%2C%20M.%2C%20Niculescu%2C%20D.%2C%20Cohen-Tannoudji%2C%20M.%2C%20Rebsam%2C%20A.%2C%20Loulier%2C%20K.%2C%20Nedelec%2C%20S.%2C%20Tozer%2C%20S.%2C%20%26amp%3B%20Livet%2C%20J.%20%282020%29.%20Direct%20Readout%20of%20Neural%20Stem%20Cell%20Transgenesis%20with%20an%20Integration-Coupled%20Gene%20Expression%20Switch.%20%3Ci%3ENeuron%3C%5C%2Fi%3E%2C%20%3Ci%3E107%3C%5C%2Fi%3E%284%29%2C%20617-630.e6.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.neuron.2020.05.038%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.neuron.2020.05.038%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Direct%20Readout%20of%20Neural%20Stem%20Cell%20Transgenesis%20with%20an%20Integration-Coupled%20Gene%20Expression%20Switch%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Takuma%22%2C%22lastName%22%3A%22Kumamoto%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Franck%22%2C%22lastName%22%3A%22Maurinot%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rapha%5Cu00eblle%22%2C%22lastName%22%3A%22Barry-Martinet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C%5Cu00e9lia%22%2C%22lastName%22%3A%22Vaslin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sandrine%22%2C%22lastName%22%3A%22Vandormael-Pournin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Micka%5Cu00ebl%22%2C%22lastName%22%3A%22Le%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marion%22%2C%22lastName%22%3A%22Lerat%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Dragos%22%2C%22lastName%22%3A%22Niculescu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michel%22%2C%22lastName%22%3A%22Cohen-Tannoudji%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alexandra%22%2C%22lastName%22%3A%22Rebsam%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Karine%22%2C%22lastName%22%3A%22Loulier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phane%22%2C%22lastName%22%3A%22Nedelec%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Samuel%22%2C%22lastName%22%3A%22Tozer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean%22%2C%22lastName%22%3A%22Livet%22%7D%5D%2C%22abstractNote%22%3A%22Stable%20genomic%20integration%20of%20exogenous%20transgenes%20is%20essential%20in%20neurodevelopmental%20and%20stem%20cell%20studies.%20Despite%20tools%20driving%20increasingly%20efficient%20genomic%20insertion%20with%20DNA%20vectors%2C%20transgenesis%20remains%20fundamentally%20hindered%20by%20the%20impossibility%20of%20distinguishing%20integrated%20from%20episomal%20transgenes.%20Here%2C%20we%20introduce%20an%20integration-coupled%20On%20genetic%20switch%2C%20iOn%2C%20which%20triggers%20gene%20expression%20upon%20incorporation%20into%20the%20host%20genome%20through%20transposition%2C%20thus%20enabling%20rapid%20and%20accurate%20identification%20of%20integration%20events%20following%20transfection%20with%20naked%20plasmids.%20In%5Cu00a0vitro%2C%20iOn%20permits%20rapid%20drug-free%20stable%20transgenesis%20of%20mouse%20and%20human%20pluripotent%20stem%20cells%20with%20multiple%20vectors.%20In%5Cu00a0vivo%2C%20we%20demonstrate%20faithful%20cell%20lineage%20tracing%2C%20assessment%20of%20regulatory%20elements%2C%20and%20mosaic%20analysis%20of%20gene%20function%20in%20somatic%20transgenesis%20experiments%20that%20reveal%20neural%20progenitor%20potentialities%20and%20interaction.%20These%20results%20establish%20iOn%20as%20a%20universally%20applicable%20strategy%20to%20accelerate%20and%20simplify%20genetic%20engineering%20in%20cultured%20systems%20and%20model%20organisms%20by%20conditioning%20transgene%20activation%20to%20genomic%20integration.%22%2C%22date%22%3A%222020-08-19%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.neuron.2020.05.038%22%2C%22ISSN%22%3A%221097-4199%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A47Z%22%7D%7D%2C%7B%22key%22%3A%22KQR7QR5U%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Darrigrand%20et%20al.%22%2C%22parsedDate%22%3A%222020-02-27%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EDarrigrand%2C%20J.-F.%2C%20Valente%2C%20M.%2C%20Comai%2C%20G.%2C%20Martinez%2C%20P.%2C%20Petit%2C%20M.%2C%20Nishinakamura%2C%20R.%2C%20Osorio%2C%20D.%20S.%2C%20Renault%2C%20G.%2C%20Marchiol%2C%20C.%2C%20Ribes%2C%20V.%2C%20%26amp%3B%20Cadot%2C%20B.%20%282020%29.%20Dullard-mediated%20Smad1%5C%2F5%5C%2F8%20inhibition%20controls%20mouse%20cardiac%20neural%20crest%20cells%20condensation%20and%20outflow%20tract%20septation.%20%3Ci%3EELife%3C%5C%2Fi%3E%2C%20%3Ci%3E9%3C%5C%2Fi%3E%2C%20e50325.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.50325%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.50325%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Dullard-mediated%20Smad1%5C%2F5%5C%2F8%20inhibition%20controls%20mouse%20cardiac%20neural%20crest%20cells%20condensation%20and%20outflow%20tract%20septation%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Fran%5Cu00e7ois%22%2C%22lastName%22%3A%22Darrigrand%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mariana%22%2C%22lastName%22%3A%22Valente%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Glenda%22%2C%22lastName%22%3A%22Comai%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pauline%22%2C%22lastName%22%3A%22Martinez%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Maxime%22%2C%22lastName%22%3A%22Petit%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ryuichi%22%2C%22lastName%22%3A%22Nishinakamura%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Daniel%20S.%22%2C%22lastName%22%3A%22Osorio%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gilles%22%2C%22lastName%22%3A%22Renault%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Carmen%22%2C%22lastName%22%3A%22Marchiol%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Ribes%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bruno%22%2C%22lastName%22%3A%22Cadot%22%7D%5D%2C%22abstractNote%22%3A%22The%20establishment%20of%20separated%20pulmonary%20and%20systemic%20circulation%20in%20vertebrates%2C%20via%20cardiac%20outflow%20tract%20%28OFT%29%20septation%2C%20is%20a%20sensitive%20developmental%20process%20accounting%20for%2010%25%20of%20all%20congenital%20anomalies.%20Neural%20Crest%20Cells%20%28NCC%29%20colonising%20the%20heart%20condensate%20along%20the%20primitive%20endocardial%20tube%20and%20force%20its%20scission%20into%20two%20tubes.%20Here%2C%20we%20show%20that%20NCC%20aggregation%20progressively%20decreases%20along%20the%20OFT%20distal-proximal%20axis%20following%20a%20BMP%20signalling%20gradient.%20Dullard%2C%20a%20nuclear%20phosphatase%2C%20tunes%20the%20BMP%20gradient%20amplitude%20and%20prevents%20NCC%20premature%20condensation.%20Dullard%20maintains%20transcriptional%20programs%20providing%20NCC%20with%20mesenchymal%20traits.%20It%20attenuates%20the%20expression%20of%20the%20aggregation%20factor%20Sema3c%20and%20conversely%20promotes%20that%20of%20the%20epithelial-mesenchymal%20transition%20driver%20Twist1.%20Altogether%2C%20Dullard-mediated%20fine-tuning%20of%20BMP%20signalling%20ensures%20the%20timed%20and%20progressive%20zipper-like%20closure%20of%20the%20OFT%20by%20the%20NCC%20and%20prevents%20the%20formation%20of%20a%20heart%20carrying%20the%20congenital%20abnormalities%20defining%20the%20tetralogy%20of%20Fallot.%22%2C%22date%22%3A%222020-02-27%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.7554%5C%2FeLife.50325%22%2C%22ISSN%22%3A%222050-084X%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A48Z%22%7D%7D%2C%7B%22key%22%3A%22SWPJ8ZNP%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Yvernogeau%20et%20al.%22%2C%22parsedDate%22%3A%222019-11%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EYvernogeau%2C%20L.%2C%20Gautier%2C%20R.%2C%20Petit%2C%20L.%2C%20Khoury%2C%20H.%2C%20Relaix%2C%20F.%2C%20Ribes%2C%20V.%2C%20Sang%2C%20H.%2C%20Charbord%2C%20P.%2C%20Souyri%2C%20M.%2C%20Robin%2C%20C.%2C%20%26amp%3B%20Jaffredo%2C%20T.%20%282019%29.%20In%20vivo%20generation%20of%20haematopoietic%20stem%5C%2Fprogenitor%20cells%20from%20bone%20marrow-derived%20haemogenic%20endothelium.%20%3Ci%3ENature%20Cell%20Biology%3C%5C%2Fi%3E%2C%20%3Ci%3E21%3C%5C%2Fi%3E%2811%29%2C%201334%26%23×2013%3B1345.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41556-019-0410-6%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41556-019-0410-6%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22In%20vivo%20generation%20of%20haematopoietic%20stem%5C%2Fprogenitor%20cells%20from%20bone%20marrow-derived%20haemogenic%20endothelium%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laurent%22%2C%22lastName%22%3A%22Yvernogeau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rodolphe%22%2C%22lastName%22%3A%22Gautier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laurence%22%2C%22lastName%22%3A%22Petit%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hanane%22%2C%22lastName%22%3A%22Khoury%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fr%5Cu00e9d%5Cu00e9ric%22%2C%22lastName%22%3A%22Relaix%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Ribes%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Helen%22%2C%22lastName%22%3A%22Sang%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pierre%22%2C%22lastName%22%3A%22Charbord%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mich%5Cu00e8le%22%2C%22lastName%22%3A%22Souyri%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Catherine%22%2C%22lastName%22%3A%22Robin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thierry%22%2C%22lastName%22%3A%22Jaffredo%22%7D%5D%2C%22abstractNote%22%3A%22It%20is%20well%20established%20that%20haematopoietic%20stem%20and%20progenitor%20cells%20%28HSPCs%29%20are%20generated%20from%20a%20transient%20subset%20of%20specialized%20endothelial%20cells%20termed%20haemogenic%2C%20present%20in%20the%20yolk%20sac%2C%20placenta%20and%20aorta%2C%20through%20an%20endothelial-to-haematopoietic%20transition%20%28EHT%29.%20HSPC%20generation%20via%20EHT%20is%20thought%20to%20be%20restricted%20to%20the%20early%20stages%20of%20development.%20By%20using%20experimental%20embryology%20and%20genetic%20approaches%20in%20birds%20and%20mice%2C%20respectively%2C%20we%20document%20here%20the%20discovery%20of%20a%20bone%20marrow%20haemogenic%20endothelium%20in%20the%20late%20fetus%5C%2Fyoung%20adult.%20These%20cells%20are%20capable%20of%20de%20novo%20producing%20a%20cohort%20of%20HSPCs%20in%20situ%20that%20harbour%20a%20very%20specific%20molecular%20signature%20close%20to%20that%20of%20aortic%20endothelial%20cells%20undergoing%20EHT%20or%20their%20immediate%20progenies%2C%20i.e.%2C%20recently%20emerged%20HSPCs.%20Taken%20together%2C%20our%20results%20reveal%20that%20HSPCs%20can%20be%20generated%20de%20novo%20past%20embryonic%20stages.%20Understanding%20the%20molecular%20events%20controlling%20this%20production%20will%20be%20critical%20for%20devising%20innovative%20therapies.%22%2C%22date%22%3A%222019-11%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41556-019-0410-6%22%2C%22ISSN%22%3A%221476-4679%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A48Z%22%7D%7D%2C%7B%22key%22%3A%22VVE3K6R4%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Duval%20et%20al.%22%2C%22parsedDate%22%3A%222019-07-25%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EDuval%2C%20N.%2C%20Vaslin%2C%20C.%2C%20Barata%2C%20T.%20C.%2C%20Frarma%2C%20Y.%2C%20Contremoulins%2C%20V.%2C%20Baudin%2C%20X.%2C%20Nedelec%2C%20S.%2C%20%26amp%3B%20Ribes%2C%20V.%20C.%20%282019%29.%20BMP4%20patterns%20Smad%20activity%20and%20generates%20stereotyped%20cell%20fate%20organization%20in%20spinal%20organoids.%20%3Ci%3EDevelopment%20%28Cambridge%2C%20England%29%3C%5C%2Fi%3E%2C%20%3Ci%3E146%3C%5C%2Fi%3E%2814%29%2C%20dev175430.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1242%5C%2Fdev.175430%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1242%5C%2Fdev.175430%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22BMP4%20patterns%20Smad%20activity%20and%20generates%20stereotyped%20cell%20fate%20organization%20in%20spinal%20organoids%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nathalie%22%2C%22lastName%22%3A%22Duval%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C%5Cu00e9lia%22%2C%22lastName%22%3A%22Vaslin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Tiago%20C.%22%2C%22lastName%22%3A%22Barata%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Youcef%22%2C%22lastName%22%3A%22Frarma%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vincent%22%2C%22lastName%22%3A%22Contremoulins%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Xavier%22%2C%22lastName%22%3A%22Baudin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phane%22%2C%22lastName%22%3A%22Nedelec%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%20C.%22%2C%22lastName%22%3A%22Ribes%22%7D%5D%2C%22abstractNote%22%3A%22Bone%20morphogenetic%20proteins%20%28BMPs%29%20are%20secreted%20regulators%20of%20cell%20fate%20in%20several%20developing%20tissues.%20In%20the%20embryonic%20spinal%20cord%2C%20they%20control%20the%20emergence%20of%20the%20neural%20crest%2C%20roof%20plate%20and%20distinct%20subsets%20of%20dorsal%20interneurons.%20Although%20a%20gradient%20of%20BMP%20activity%20has%20been%20proposed%20to%20determine%20cell%20type%20identity%20in%20vivo%2C%20whether%20this%20is%20sufficient%20for%20pattern%20formation%20in%20vitro%20is%20unclear.%20Here%2C%20we%20demonstrate%20that%20exposure%20to%20BMP4%20initiates%20distinct%20spatial%20dynamics%20of%20BMP%20signalling%20within%20the%20self-emerging%20epithelia%20of%20both%20mouse%20and%20human%20pluripotent%20stem%20cell-derived%20spinal%20organoids.%20The%20pattern%20of%20BMP%20signalling%20results%20in%20the%20stereotyped%20spatial%20arrangement%20of%20dorsal%20neural%20tube%20cell%20types%2C%20and%20concentration%2C%20timing%20and%20duration%20of%20BMP4%20exposure%20modulate%20these%20patterns.%20Moreover%2C%20differences%20in%20the%20duration%20of%20competence%20time-windows%20between%20mouse%20and%20human%20account%20for%20the%20species-specific%20tempo%20of%20neural%20differentiation.%20Together%2C%20this%20study%20describes%20efficient%20methods%20for%20generating%20patterned%20subsets%20of%20dorsal%20interneurons%20in%20spinal%20organoids%20and%20supports%20the%20conclusion%20that%20graded%20BMP%20activity%20orchestrates%20the%20spatial%20organization%20of%20the%20dorsal%20neural%20tube%20cellular%20diversity%20in%20mouse%20and%20human.%22%2C%22date%22%3A%222019-07-25%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1242%5C%2Fdev.175430%22%2C%22ISSN%22%3A%221477-9129%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A47Z%22%7D%7D%2C%7B%22key%22%3A%22EFD4DE86%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Gerber%20et%20al.%22%2C%22parsedDate%22%3A%222019-02-20%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EGerber%2C%20V.%2C%20Yang%2C%20L.%2C%20Takamiya%2C%20M.%2C%20Ribes%2C%20V.%2C%20Gourain%2C%20V.%2C%20Peravali%2C%20R.%2C%20Stegmaier%2C%20J.%2C%20Mikut%2C%20R.%2C%20Reischl%2C%20M.%2C%20Ferg%2C%20M.%2C%20Rastegar%2C%20S.%2C%20%26amp%3B%20Str%26%23xE4%3Bhle%2C%20U.%20%282019%29.%20The%20HMG%20box%20transcription%20factors%20Sox1a%20and%20Sox1b%20specify%20a%20new%20class%20of%20glycinergic%20interneuron%20in%20the%20spinal%20cord%20of%20zebrafish%20embryos.%20%3Ci%3EDevelopment%20%28Cambridge%2C%20England%29%3C%5C%2Fi%3E%2C%20%3Ci%3E146%3C%5C%2Fi%3E%284%29%2C%20dev172510.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1242%5C%2Fdev.172510%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1242%5C%2Fdev.172510%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20HMG%20box%20transcription%20factors%20Sox1a%20and%20Sox1b%20specify%20a%20new%20class%20of%20glycinergic%20interneuron%20in%20the%20spinal%20cord%20of%20zebrafish%20embryos%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Gerber%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lixin%22%2C%22lastName%22%3A%22Yang%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Masanari%22%2C%22lastName%22%3A%22Takamiya%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Ribes%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Victor%22%2C%22lastName%22%3A%22Gourain%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ravindra%22%2C%22lastName%22%3A%22Peravali%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Johannes%22%2C%22lastName%22%3A%22Stegmaier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ralf%22%2C%22lastName%22%3A%22Mikut%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Markus%22%2C%22lastName%22%3A%22Reischl%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marco%22%2C%22lastName%22%3A%22Ferg%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sepand%22%2C%22lastName%22%3A%22Rastegar%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Uwe%22%2C%22lastName%22%3A%22Str%5Cu00e4hle%22%7D%5D%2C%22abstractNote%22%3A%22Specification%20of%20neurons%20in%20the%20spinal%20cord%20relies%20on%20extrinsic%20and%20intrinsic%20signals%2C%20which%20in%20turn%20are%20interpreted%20by%20expression%20of%20transcription%20factors.%20V2%20interneurons%20develop%20from%20the%20ventral%20aspects%20of%20the%20spinal%20cord.%20We%20report%20here%20a%20novel%20neuronal%20V2%20subtype%2C%20named%20V2s%2C%20in%20zebrafish%20embryos.%20Formation%20of%20these%20neurons%20depends%20on%20the%20transcription%20factors%20sox1a%20and%20sox1b.%20They%20develop%20from%20common%20gata2a-%20and%20gata3-dependent%20precursors%20co-expressing%20markers%20of%20V2b%20and%20V2s%20interneurons.%20Chemical%20blockage%20of%20Notch%20signalling%20causes%20a%20decrease%20in%20V2s%20and%20an%20increase%20in%20V2b%20cells.%20Our%20results%20are%20consistent%20with%20the%20existence%20of%20at%20least%20two%20types%20of%20precursor%20arranged%20in%20a%20hierarchical%20manner%20in%20the%20V2%20domain.%20V2s%20neurons%20grow%20long%20ipsilateral%20descending%20axonal%20projections%20with%20a%20short%20branch%20at%20the%20ventral%20midline.%20They%20acquire%20a%20glycinergic%20neurotransmitter%20type%20during%20the%20second%20day%20of%20development.%20Unilateral%20ablation%20of%20V2s%20interneurons%20causes%20a%20delay%20in%20touch-provoked%20escape%20behaviour%2C%20suggesting%20that%20V2s%20interneurons%20are%20involved%20in%20fast%20motor%20responses.%22%2C%22date%22%3A%222019-02-20%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1242%5C%2Fdev.172510%22%2C%22ISSN%22%3A%221477-9129%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A48Z%22%7D%7D%2C%7B%22key%22%3A%22ZNSWE8IB%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Gard%20et%20al.%22%2C%22parsedDate%22%3A%222017-12-01%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EGard%2C%20C.%2C%20Gonzalez%20Curto%2C%20G.%2C%20Frarma%2C%20Y.%20E.-M.%2C%20Chollet%2C%20E.%2C%20Duval%2C%20N.%2C%20Auzi%26%23xE9%3B%2C%20V.%2C%20Aurad%26%23xE9%3B%2C%20F.%2C%20Vigier%2C%20L.%2C%20Relaix%2C%20F.%2C%20Pierani%2C%20A.%2C%20Causeret%2C%20F.%2C%20%26amp%3B%20Ribes%2C%20V.%20%282017%29.%20Pax3-%20and%20Pax7-mediated%20Dbx1%20regulation%20orchestrates%20the%20patterning%20of%20intermediate%20spinal%20interneurons.%20%3Ci%3EDevelopmental%20Biology%3C%5C%2Fi%3E%2C%20%3Ci%3E432%3C%5C%2Fi%3E%281%29%2C%2024%26%23×2013%3B33.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.ydbio.2017.06.014%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.ydbio.2017.06.014%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Pax3-%20and%20Pax7-mediated%20Dbx1%20regulation%20orchestrates%20the%20patterning%20of%20intermediate%20spinal%20interneurons%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Chris%22%2C%22lastName%22%3A%22Gard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gloria%22%2C%22lastName%22%3A%22Gonzalez%20Curto%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Youcef%20El-Mokhtar%22%2C%22lastName%22%3A%22Frarma%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elodie%22%2C%22lastName%22%3A%22Chollet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nathalie%22%2C%22lastName%22%3A%22Duval%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Valentine%22%2C%22lastName%22%3A%22Auzi%5Cu00e9%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fr%5Cu00e9d%5Cu00e9ric%22%2C%22lastName%22%3A%22Aurad%5Cu00e9%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lisa%22%2C%22lastName%22%3A%22Vigier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fr%5Cu00e9d%5Cu00e9ric%22%2C%22lastName%22%3A%22Relaix%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alessandra%22%2C%22lastName%22%3A%22Pierani%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Fr%5Cu00e9d%5Cu00e9ric%22%2C%22lastName%22%3A%22Causeret%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vanessa%22%2C%22lastName%22%3A%22Ribes%22%7D%5D%2C%22abstractNote%22%3A%22Transcription%20factors%20are%20key%20orchestrators%20of%20the%20emergence%20of%20neuronal%20diversity%20within%20the%20developing%20spinal%20cord.%20As%20such%2C%20the%20two%20paralogous%20proteins%20Pax3%20and%20Pax7%20regulate%20the%20specification%20of%20progenitor%20cells%20within%20the%20intermediate%20neural%20tube%2C%20by%20defining%20a%20neat%20segregation%20between%20those%20fated%20to%20form%20motor%20circuits%20and%20those%20involved%20in%20the%20integration%20of%20sensory%20inputs.%20To%20attain%20insights%20into%20the%20molecular%20means%20by%20which%20they%20control%20this%20process%2C%20we%20have%20performed%20detailed%20phenotypic%20analyses%20of%20the%20intermediate%20spinal%20interneurons%20%28IN%29%2C%20namely%20the%20dI6%2C%20V0D%2C%20V0VCG%20and%20V1%20populations%20in%20compound%20null%20mutants%20for%20Pax3%20and%20Pax7.%20This%20has%20revealed%20that%20the%20levels%20of%20Pax3%5C%2F7%20proteins%20determine%20both%20the%20dorso-ventral%20extent%20and%20the%20number%20of%20cells%20produced%20in%20each%20subpopulation%3B%20with%20increasing%20levels%20leading%20to%20the%20dorsalisation%20of%20their%20fate.%20Furthermore%2C%20thanks%20to%20the%20examination%20of%20mutants%20in%20which%20Pax3%20transcriptional%20activity%20is%20skewed%20either%20towards%20repression%20or%20activation%2C%20we%20demonstrate%20that%20this%20cell%20diversification%20process%20is%20mainly%20dictated%20by%20Pax3%5C%2F7%20ability%20to%20repress%20gene%20expression.%20Consistently%2C%20we%20show%20that%20Pax3%20and%20Pax7%20inhibit%20the%20expression%20of%20Dbx1%20and%20of%20its%20repressor%20Prdm12%2C%20fate%20determinants%20of%20the%20V0%20and%20V1%20interneurons%2C%20respectively.%20Notably%2C%20we%20provide%20evidence%20for%20the%20activity%20of%20several%20cis-regulatory%20modules%20of%20Dbx1%20to%20be%20sensitive%20to%20Pax3%20and%20Pax7%20transcriptional%20activity%20levels.%20Altogether%2C%20our%20study%20provides%20insights%20into%20how%20the%20redundancy%20within%20a%20TF%20family%2C%20together%20with%20discrete%20dynamics%20of%20expression%20profiles%20of%20each%20member%2C%20are%20exploited%20to%20generate%20cellular%20diversity.%20Furthermore%2C%20our%20data%20supports%20the%20model%20whereby%20cell%20fate%20choices%20in%20the%20neural%20tube%20do%20not%20rely%20on%20binary%20decisions%20but%20rather%20on%20inhibition%20of%20multiple%20alternative%20fates.%22%2C%22date%22%3A%222017-12-01%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.ydbio.2017.06.014%22%2C%22ISSN%22%3A%221095-564X%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A48Z%22%7D%7D%2C%7B%22key%22%3A%22JCKGD53A%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Maury%20et%20al.%22%2C%22parsedDate%22%3A%222015-01%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EMaury%2C%20Y.%2C%20C%26%23xF4%3Bme%2C%20J.%2C%20Piskorowski%2C%20R.%20A.%2C%20Salah-Mohellibi%2C%20N.%2C%20Chevaleyre%2C%20V.%2C%20Peschanski%2C%20M.%2C%20Martinat%2C%20C.%2C%20%26amp%3B%20Nedelec%2C%20S.%20%282015%29.%20Combinatorial%20analysis%20of%20developmental%20cues%20efficiently%20converts%20human%20pluripotent%20stem%20cells%20into%20multiple%20neuronal%20subtypes.%20%3Ci%3ENature%20Biotechnology%3C%5C%2Fi%3E%2C%20%3Ci%3E33%3C%5C%2Fi%3E%281%29%2C%2089%26%23×2013%3B96.%20%3Ca%20class%3D%27zp-DOIURL%27%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fnbt.3049%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fnbt.3049%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Combinatorial%20analysis%20of%20developmental%20cues%20efficiently%20converts%20human%20pluripotent%20stem%20cells%20into%20multiple%20neuronal%20subtypes%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yves%22%2C%22lastName%22%3A%22Maury%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julien%22%2C%22lastName%22%3A%22C%5Cu00f4me%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rebecca%20A.%22%2C%22lastName%22%3A%22Piskorowski%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nouzha%22%2C%22lastName%22%3A%22Salah-Mohellibi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vivien%22%2C%22lastName%22%3A%22Chevaleyre%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marc%22%2C%22lastName%22%3A%22Peschanski%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22C%5Cu00e9cile%22%2C%22lastName%22%3A%22Martinat%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phane%22%2C%22lastName%22%3A%22Nedelec%22%7D%5D%2C%22abstractNote%22%3A%22Specification%20of%20cell%20identity%20during%20development%20depends%20on%20exposure%20of%20cells%20to%20sequences%20of%20extrinsic%20cues%20delivered%20at%20precise%20times%20and%20concentrations.%20Identification%20of%20combinations%20of%20patterning%20molecules%20that%20control%20cell%20fate%20is%20essential%20for%20the%20effective%20use%20of%20human%20pluripotent%20stem%20cells%20%28hPSCs%29%20for%20basic%20and%20translational%20studies.%20Here%20we%20describe%20a%20scalable%2C%20automated%20approach%20to%20systematically%20test%20the%20combinatorial%20actions%20of%20small%20molecules%20for%20the%20targeted%20differentiation%20of%20hPSCs.%20Applied%20to%20the%20generation%20of%20neuronal%20subtypes%2C%20this%20analysis%20revealed%20an%20unappreciated%20role%20for%20canonical%20Wnt%20signaling%20in%20specifying%20motor%20neuron%20diversity%20from%20hPSCs%20and%20allowed%20us%20to%20define%20rapid%20%2814%20days%29%2C%20efficient%20procedures%20to%20generate%20spinal%20and%20cranial%20motor%20neurons%20as%20well%20as%20spinal%20interneurons%20and%20sensory%20neurons.%20Our%20systematic%20approach%20to%20improving%20hPSC-targeted%20differentiation%20should%20facilitate%20disease%20modeling%20studies%20and%20drug%20screening%20assays.%22%2C%22date%22%3A%222015-01%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1038%5C%2Fnbt.3049%22%2C%22ISSN%22%3A%221546-1696%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%22MIYUW3RH%22%5D%2C%22dateModified%22%3A%222025-02-11T10%3A55%3A47Z%22%7D%7D%5D%7D
Rondon, R., Hezez, T., Albert, J. R., Drayton-Libotte, B., Gonzalez-Curto, G., Relaix, F., Dugast-Darzacq, C., Gilardi-Hebenstreit, P., & Ribes, V. (2025).
Dual PAX3/7 transcriptional activities spatially encode spinal cell fates through distinct gene networks. bioRxiv.
https://doi.org/10.1101/2025.03.11.642668
Huchede, P., Meyer, S., Berthelot, C., Hamadou, M., Bertrand-Chapel, A., Rakotomalala, A., Manceau, L., Tomine, J., Lespinasse, N., Lewandowski, P., Cordier-Bussat, M., Broutier, L., Dutour, A., Rochet, I., Blay, J.-Y., Degletagne, C., Attignon, V., Montero-Carcaboso, A., Le Grand, M., … Castets, M. (2024). BMP2 and BMP7 cooperate with H3.3K27M to promote quiescence and invasiveness in pediatric diffuse midline gliomas.
ELife,
12, RP91313.
https://doi.org/10.7554/eLife.91313
Rodríguez-Martín, M., Báez-Flores, J., Ribes, V., Isidoro-García, M., Lacal, J., & Prieto-Matos, P. (2024). Non-Mammalian Models for Understanding Neurological Defects in RASopathies.
Biomedicines,
12(4), 841.
https://doi.org/10.3390/biomedicines12040841
Mirdass, C., Catala, M., Bocel, M., Nedelec, S., & Ribes, V. (2023). Stem cell-derived models of spinal neurulation.
Emerging Topics in Life Sciences,
7(4), 423–437.
https://doi.org/10.1042/ETLS20230087
Gribaudo, S., Robert, R., van Sambeek, B., Mirdass, C., Lyubimova, A., Bouhali, K., Ferent, J., Morin, X., van Oudenaarden, A., & Nedelec, S. (2023). Self-organizing models of human trunk organogenesis recapitulate spinal cord and spine co-morphogenesis.
Nature Biotechnology.
https://doi.org/10.1038/s41587-023-01956-9
Liau, E. S., Jin, S., Chen, Y.-C., Liu, W.-S., Calon, M., Nedelec, S., Nie, Q., & Chen, J.-A. (2023). Single-cell transcriptomic analysis reveals diversity within mammalian spinal motor neurons.
Nature Communications,
14(1), 46.
https://doi.org/10.1038/s41467-022-35574-x
Manceau, L., Richard Albert, J., Lollini, P.-L., Greenberg, M. V. C., Gilardi-Hebenstreit, P., & Ribes, V. (2022). Divergent transcriptional and transforming properties of PAX3-FOXO1 and PAX7-FOXO1 paralogs.
PLoS Genetics,
18(5), e1009782.
https://doi.org/10.1371/journal.pgen.1009782
Mouilleau, V., Vaslin, C., Robert, R., Gribaudo, S., Nicolas, N., Jarrige, M., Terray, A., Lesueur, L., Mathis, M. W., Croft, G., Daynac, M., Rouiller-Fabre, V., Wichterle, H., Ribes, V., Martinat, C., & Nedelec, S. (2021). Dynamic extrinsic pacing of the HOX clock in human axial progenitors controls motor neuron subtype specification.
Development (Cambridge, England),
148(6), dev194514.
https://doi.org/10.1242/dev.194514
Nedelec, S., & Martinez-Arias, A. (2021). In vitro models of spinal motor circuit’s development in mammals: achievements and challenges.
Current Opinion in Neurobiology,
66, 240–249.
https://doi.org/10.1016/j.conb.2020.12.002
Gonzalez Curto, G., Der Vartanian, A., Frarma, Y. E.-M., Manceau, L., Baldi, L., Prisco, S., Elarouci, N., Causeret, F., Korenkov, D., Rigolet, M., Aurade, F., De Reynies, A., Contremoulins, V., Relaix, F., Faklaris, O., Briscoe, J., Gilardi-Hebenstreit, P., & Ribes, V. (2020). The PAX-FOXO1s trigger fast trans-differentiation of chick embryonic neural cells into alveolar rhabdomyosarcoma with tissue invasive properties limited by S phase entry inhibition.
PLoS Genetics,
16(11), e1009164.
https://doi.org/10.1371/journal.pgen.1009164
Kumamoto, T., Maurinot, F., Barry-Martinet, R., Vaslin, C., Vandormael-Pournin, S., Le, M., Lerat, M., Niculescu, D., Cohen-Tannoudji, M., Rebsam, A., Loulier, K., Nedelec, S., Tozer, S., & Livet, J. (2020). Direct Readout of Neural Stem Cell Transgenesis with an Integration-Coupled Gene Expression Switch.
Neuron,
107(4), 617-630.e6.
https://doi.org/10.1016/j.neuron.2020.05.038
Darrigrand, J.-F., Valente, M., Comai, G., Martinez, P., Petit, M., Nishinakamura, R., Osorio, D. S., Renault, G., Marchiol, C., Ribes, V., & Cadot, B. (2020). Dullard-mediated Smad1/5/8 inhibition controls mouse cardiac neural crest cells condensation and outflow tract septation.
ELife,
9, e50325.
https://doi.org/10.7554/eLife.50325
Yvernogeau, L., Gautier, R., Petit, L., Khoury, H., Relaix, F., Ribes, V., Sang, H., Charbord, P., Souyri, M., Robin, C., & Jaffredo, T. (2019). In vivo generation of haematopoietic stem/progenitor cells from bone marrow-derived haemogenic endothelium.
Nature Cell Biology,
21(11), 1334–1345.
https://doi.org/10.1038/s41556-019-0410-6
Duval, N., Vaslin, C., Barata, T. C., Frarma, Y., Contremoulins, V., Baudin, X., Nedelec, S., & Ribes, V. C. (2019). BMP4 patterns Smad activity and generates stereotyped cell fate organization in spinal organoids.
Development (Cambridge, England),
146(14), dev175430.
https://doi.org/10.1242/dev.175430
Gerber, V., Yang, L., Takamiya, M., Ribes, V., Gourain, V., Peravali, R., Stegmaier, J., Mikut, R., Reischl, M., Ferg, M., Rastegar, S., & Strähle, U. (2019). The HMG box transcription factors Sox1a and Sox1b specify a new class of glycinergic interneuron in the spinal cord of zebrafish embryos.
Development (Cambridge, England),
146(4), dev172510.
https://doi.org/10.1242/dev.172510
Gard, C., Gonzalez Curto, G., Frarma, Y. E.-M., Chollet, E., Duval, N., Auzié, V., Auradé, F., Vigier, L., Relaix, F., Pierani, A., Causeret, F., & Ribes, V. (2017). Pax3- and Pax7-mediated Dbx1 regulation orchestrates the patterning of intermediate spinal interneurons.
Developmental Biology,
432(1), 24–33.
https://doi.org/10.1016/j.ydbio.2017.06.014
Maury, Y., Côme, J., Piskorowski, R. A., Salah-Mohellibi, N., Chevaleyre, V., Peschanski, M., Martinat, C., & Nedelec, S. (2015). Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes.
Nature Biotechnology,
33(1), 89–96.
https://doi.org/10.1038/nbt.3049
Thèses
- Maeliss Calon, «Etude des bases moléculaires et cellulaires de la vulnérabilité des neurones moteurs dans l’amyotrophie spinale distale des membres inférieurs», soutenue le 26 Septembre 2024
- Rémi Robert, «Etude des mécanismes contrôlant l’expression des gènes HOX et implications pour la génération in vitro de tissus humains», soutenue le 22 Septembre 2023
- Line Manceau, «Etude des mécanismes moléculaires et cellulaires par lesquels les facteurs de transcription paralogues PAX3-FOXO1 et PAX7-FOXO1 exercent leur activité oncogénique», soutenue le 24 Septembre 2021
- Célia Vaslin, «Etude des mécanismes de signalisation cellulaire contrôlant la diversification neuronale dans la moelle épinière», soutenue le 26 Mars 2021
- Vincent Mouilleau, «Etude des mécanismes du développement de la moelle épinière humaine par la mise en place de modèles in vitro dérivés de cellules souches pluripotentes humaines», soutenue 26 Novembre 2019
Collaborations
- Nicolas Borghi (IJM)
- Valérie Doye (IJM)
- Alexandre Baffet (Institut Curie, France)
- Nadia Bahi-Buisson (Institut Imagine, France)
- Bertrand Bénazéraf (Centre de Biologie Intégrative, CBI, France)
- Jésus Lacal (University of Salamanca, Spain)
- Marie Castets (Centre de Recherche en Cancérologie de Lyon, CRCL, Lyon)
- Valérie Dupé (Institut Génétique & Développement de Rennes, IGDR, Rennes)
- Fiona Francis (Institut du Fer à Moulin, IFM, France)
- Eddy Pasquier (Centre de Recherche en Cancérologie de Marseille, CRCM, Marseille)
- Alessandra Pierani (Institut Imagine, France)
- Benoit Sorre (Institut Curie, France)
Financements
- INSERM
- Ligue Nationale Contre le Cancer
- Agence Nationale de la Recherche
- AFM – Téléthon
- Fondation pour la Recherche Médicale
- L’association WonderAugustine
- Financement “Emergence en recherche” (Université Paris Cité)
Actualité
Emploi
Postes Post-Doctoraux
Nous recherchons des candidat(e)s post-doctorant(e)s intéressé(e)s par l’étude de la régulation transcriptionnelle des décisions de destin cellulaire, avec une expertise en culture de cellules iPSC ou en bioinformatique. Les candidat(e)s doivent envoyer leur CV, une lettre de motivation et les coordonnées de 2 à 3 référents à l’adresse suivante : stephane.nedelec@ijm.fr.
Étudiants en Master
Nous recherchons actuellement un(e) étudiant(e) de Master 2, intéressé(e) par la régulation des états de la chromatine pendant le développement neural (stephane.nedelec@ijm.fr) ou l’émergence du rhabdomyosarcome (vanessa.ribes@ijm.fr).
Recrutement international pour le projet de thèse « Modelling OPCs diversity of the hindbrain and their lineage derail during DMG oncogenesis using brain organoids »: Candidatez avant le 14 mars 2025

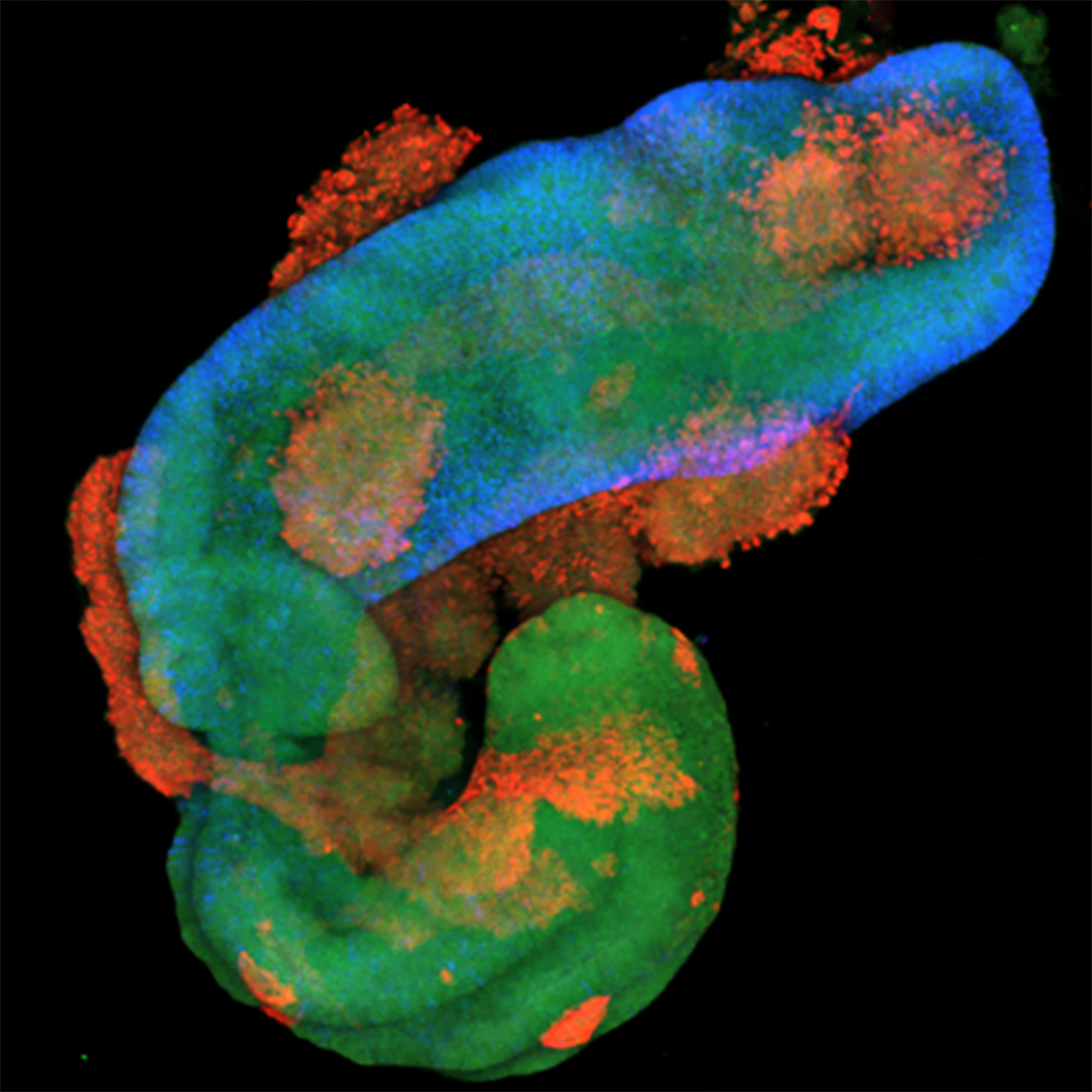


 Stephane NEDELEC, Chercheur, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Stephane NEDELEC, Chercheur, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B Vanessa RIBES, Chercheur, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Vanessa RIBES, Chercheur, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B Kenza CHERIET, Doctorante, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Kenza CHERIET, Doctorante, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B Claire DUGAST, Chercheur, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Claire DUGAST, Chercheur, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B Pascale GILARDI HEBENSTREIT, Chercheur, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Pascale GILARDI HEBENSTREIT, Chercheur, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B Marine GRISON, Ingénieure en biologie, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Marine GRISON, Ingénieure en biologie, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B Grace HENSTONE, Doctorante, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Grace HENSTONE, Doctorante, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B Theaud HEZEZ, Doctorant, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Theaud HEZEZ, Doctorant, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B Helena MALEK, Doctorante, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Helena MALEK, Doctorante, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B Camil MIRDASS, Doctorant, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Camil MIRDASS, Doctorant, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B Robin RONDON, Doctorant, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B
Robin RONDON, Doctorant, RIBES/NEDELEC LAB+33 (0)1 57 27 81 93, bureau 555B

