Cycle cellulaire et développement
LIONEL PINTARD
L’équipe Cycle Cellulaire et Développement vise à acquérir de nouvelles connaissances pour décrypter la façon dont les cellules se divisent afin de mieux comprendre les mécanismes du cancer, une maladie résultant d’une division cellulaire incontrôlée.
L’équipe utilise principalement le nématode C. elegans comme système modèle et emploie une approche multidisciplinaire combinant diverses approches (biochimie, génétique, imagerie, protéomique) permettant de poser des questions à différentes échelles, de la molécule à l’organisme. Les mécanismes de régulation de la division cellulaire étant conservés entre les espèces, l’équipe étudie les paradigmes émergeant de C. elegans dans les cellules humaines.
Mots clés: Division cellulaire – Mitose – Méiose – Kinases – Rupture de l’enveloppe nucléaire – Enzymes de coupure des Microtubules – Katanin
+33 (0)1 57 27 80 89 Contact @ccdlab.bsky.social https://sites.google.com/site/pintardlab/
Notre vision
Acquérir de nouvelles connaissances pour décrypter la façon dont les cellules se divisent afin de mieux comprendre les mécanismes du cancer, une maladie résultant d’une division cellulaire incontrôlée.
Contexte
Les êtres humains sont constitués d’environ 1013 cellules correspondant à 200 types cellulaires différents. Toutes ces cellules sont générées par divisions cellulaires, à partir d’une seule cellule, l’ovocyte fécondé. Pour générer ce grand nombre de cellules et maintenir l’homéostasie des tissus, le corps humain subit jusqu’à 1016 divisions cellulaires au cours d’une vie. Lors de chaque division cellulaire, le génome doit être reproduit fidèlement et séparé de manière égale entre les cellules filles pendant la mitose. Des défauts dans ces processus peuvent avoir des conséquences dramatiques, pouvant conduire à une croissance dérégulée, typique du cancer. Malgré des progrès considérables réalisés au cours des dernières décennies, les mécanismes qui régulent la division cellulaire sont encore mal compris, en particulier au cours du développement. Ce manque de connaissances a considérablement limité le développement d’approches thérapeutiques innovantes.
Programme de recherche
Mécanismes qui contrôlent l’entrée en mitose dans l’espace et le temps
L’entrée en mitose doit être étroitement coordonné avec la réplication de l’ADN afin de préserver l’intégrité du génome. Une entrée en mitose non programmée peut conduire à une instabilité génétique. L’entrée en mitose est contrôlée par des sérine/thréonine kinases (Aurora A, Polo-like kinase, Plk1) conservées au cours de l’évolution, ainsi que par des phosphatases (PPases). La manière dont ces activités kinases sont régulées dans l’espace et le temps, et la façon dont elles coordonnent leur activité pour déclencher l’entrée en mitose au bon moment restent mal défini.
o Mécanisme d’activation des kinases mitotiques (Aurora A, Polo-like kinase)
o Rôle des kinases mitotiques dans la rupture de l’enveloppe nucléaire (NEBD)
o Rôle et régulation des kinases mitotiques dans les divisions cellulaires asynchrones
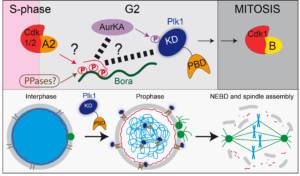
Figure 1: L’axe Bora-Aurora A-Plk1 et son rôle lors de l’entrée en mitose
La transition méiose-mitose : rôle et régulation de la Katanine
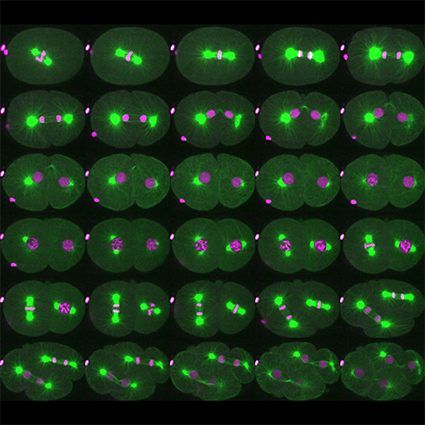
Les microtubules (MT) sont des polymères dynamiques du cytosquelette, qui jouent un rôle central dans la division cellulaire. La plupart des protéines régulatrices des MT interagissent avec l’extrémité plus ou moins des microtubules et contrôlent ainsi leur taux de polymérisation et de dépolymérisation. Une autre classe de régulateurs coupe les MT contrôlant ainsi leur taille dans la cellule. Trois enzymes conservées au cours de l’évolution capables de couper les MT ont été identifiées : Fidgetin, Spastin et Katanin. La mutation de ces enzymes est associée à divers défauts et pathologies, notamment des troubles du développement et des troubles neurodégénératifs. En outre, ces enzymes sont directement impliquées sans la division cellulaire. Cependant, le mécanisme moléculaire par lesquel ces enzymes coupent les MT reste mal compris. De même, les mécanismes mis en jeu pour contrôler l’activité de coupure dans l’espace et dans le temps restent à découvrir. Nous nous concentrons actuellement sur le décryptage du mode d’action et de la régulation de la Katanine, qui est essentielle pour l’assemblage du fuseau méiotique femelle chez C. elegans.
o Rôle de la coupure des MT dans l’assemblage du fuseau méiotique
o Mécanisme par lequel la Katanine coupe les MT
o Régulation de l’activité Katanine dans l’espace et le temps pendant le développement
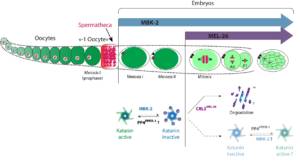 Figure 2: Role and regulation de la Katanin lors du développement de C. elegans (From Joly et al. JCB 2020).
Figure 2: Role and regulation de la Katanin lors du développement de C. elegans (From Joly et al. JCB 2020).
Les ubiquitine-ligases Cullin-RING E3-Ligases dans la division cellulaire
Les ubiquitine-ligases nucléés par les cullines (CRL pour Cullin-RING E3-ligases) représentent la plus grande famille d’ubiquitine-ligases ciblant la dégradation des principaux régulateurs du cycle cellulaire dans l’espace et le temps, contribuant ainsi à la progression ordonnée du cycle de division cellulaire. Nous cherchons à comprendre comment ces enzymes régulent la progression du cycle cellulaire dans un contexte de développement.
o CRL dans la régulation de la voie Bora-Aurora-Plk1
o CRL dans la régulation de l’activité de la Katanine
o CRL dans le maintien de l’intégrité de la réplication de l’ADN
Approches
Nous utilisons une approche multidisciplinaire comprenant la biochimie (reconstitution d’activités enzymatiques à partir de composants purifiés pour disséquer les mécanismes moléculaires), la génétique, l’imagerie des cellules vivantes, les approches protéomiques utilisant à la fois des cellules humaines et le nématode C. elegans. Les mécanismes de régulation de la division cellulaire sont conservés entre les espèces, de sorte que le paradigme émergeant de C. elegans peut être immédiatement étudié dans les cellules humaines. En outre, C. elegans offre un certain nombre d’avantages pratiques pour l’étude des voies conservées régulant la division cellulaire (Pintard & Bowerman, Genetics 2019).
Membres








Pour contacter un membre de l’équipe par mail : prenom.nom@ijm.fr

June, 2021 (c) Pintard Lab
From left to right: Anais, Griselda, Batool, Lucie, NicoT, Sylvia, Lionel, Eva, Lola, Anaelle, Emma, NicoJ
Roumbo, L., Ossareh-Nazari, B., Vigneron, S., Stefani, I., Van Hove, L., Legros, V., Chevreux, G., Lacroix, B., Castro, A., Joly, N., Lorca, T., & Pintard, L. (2025). The MAST kinase KIN-4 carries out mitotic entry functions of Greatwall in C. elegans. The EMBO Journal, 1–32. https://doi.org/10.1038/s44318-025-00364-w
Beaumale, E., Van Hove, L., Pintard, L., & Joly, N. (2024). Microtubule-binding domains in Katanin p80 subunit are essential for severing activity in C. elegans. Journal of Cell Biology, 223(4), e202308023. https://doi.org/10.1083/jcb.202308023
Nkombo Nkoula, S., Velez-Aguilera, G., Ossareh-Nazari, B., Van Hove, L., Ayuso, C., Legros, V., Chevreux, G., Thomas, L., Seydoux, G., Askjaer, P., & Pintard, L. (2023). Mechanisms of nuclear pore complex disassembly by the mitotic Polo-like kinase 1 (PLK-1) in C. elegans embryos. Science Advances, 9(29), eadf7826. https://doi.org/10.1126/sciadv.adf7826
Velez-Aguilera, G., Ossareh-Nazari, B., Van Hove, L., Joly, N., & Pintard, L. (2022). Cortical microtubule pulling forces contribute to the union of the parental genomes in the Caenorhabditis elegans zygote. ELife, 11, e75382. https://doi.org/10.7554/eLife.75382
Tavernier, N., Thomas, Y., Vigneron, S., Maisonneuve, P., Orlicky, S., Mader, P., Regmi, S. G., Van Hove, L., Levinson, N. M., Gasmi-Seabrook, G., Joly, N., Poteau, M., Velez-Aguilera, G., Gavet, O., Castro, A., Dasso, M., Lorca, T., Sicheri, F., & Pintard, L. (2021). Bora phosphorylation substitutes in trans for T-loop phosphorylation in Aurora A to promote mitotic entry. Nature Communications, 12(1), 1899. https://doi.org/10.1038/s41467-021-21922-w
Publications
2913254
GWSKYQWE
1
apa
50
date
desc
8942
https://www.ijm.fr/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3Afalse%2C%22meta%22%3A%7B%22request_last%22%3A0%2C%22request_next%22%3A0%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%22YNT9FYUT%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Joly%20and%20Pintard%22%2C%22parsedDate%22%3A%222025-11-19%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BJoly%2C%20N.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282025%29.%20Intramolecular%20regulation%20of%20the%20MT-severing%20enzyme%20Katanin%20prevents%20futile%20ATP%20hydrolysis.%20%26lt%3Bi%26gt%3BJournal%20of%20Cell%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B225%26lt%3B%5C%2Fi%26gt%3B%282%29%2C%20e202506192.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202506192%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202506192%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Intramolecular%20regulation%20of%20the%20MT-severing%20enzyme%20Katanin%20prevents%20futile%20ATP%20hydrolysis%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22Microtubule-severing%20enzymes%20are%20evolutionarily%20conserved%20AAA-ATPases%20that%20sever%20microtubules%2C%20thereby%20regulating%20diverse%20microtubule-dependent%20cellular%20processes.%20How%20these%20enzymes%20couple%20Microtubule%20binding%20with%20ATP%20hydrolysis%20to%20trigger%20microtubule-remodeling%20remains%20poorly%20understood.%20Using%20Caenorhabditiselegans%20Katanin%2C%20which%20contains%20the%20MEI-1%20catalytic%20AAA%2B%20p60%20and%20MEI-2%20p80-like%20regulatory%20subunits%2C%20we%20identify%20a%20critical%20regulatory%20role%20of%20the%20N-terminal%20domain%20of%20MEI-1%20in%20Katanin%20regulation.%20We%20demonstrate%20this%20domain%20represses%20the%20AAA%2B%20core%20in%20cis%2C%20limiting%20ATP%20hydrolysis%20and%20preventing%20interaction%20with%20tubulin%20C-terminal%20tails%20in%20the%20absence%20of%20MEI-2.%20Strikingly%2C%20MEI-1%20lacking%20its%20N%20terminus%20is%20constitutively%20active%2C%20enabling%20identification%20of%20pore%20residues%20critical%20for%20sensing%20microtubule%20C-terminal%20tails%20and%20relaying%20this%20signal%20to%20the%20AAA%2B%20core.%20These%20findings%20reveal%20how%20Katanin%20activation%20is%20coupled%20to%20microtubule%20binding%2C%20thereby%20avoiding%20futile%20ATP%20hydrolysis.%20Given%20Katanin%5Cu2019s%20evolutionary%20conservation%2C%20our%20work%20provides%20a%20mechanistic%20framework%20for%20its%20regulation%20in%20other%20organisms%2C%20with%20broader%20implications%20for%20human%20pathologies%2C%20including%20neurodegeneration%20and%20cancer.%22%2C%22date%22%3A%222025-11-19%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1083%5C%2Fjcb.202506192%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202506192%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220021-9525%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222025-11-25T15%3A20%3A51Z%22%7D%7D%2C%7B%22key%22%3A%224NIR3VJ9%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Roumbo%20et%20al.%22%2C%22parsedDate%22%3A%222025-02-17%22%2C%22numChildren%22%3A0%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BRoumbo%2C%20L.%2C%20Ossareh-Nazari%2C%20B.%2C%20Vigneron%2C%20S.%2C%20Stefani%2C%20I.%2C%20Van%20Hove%2C%20L.%2C%20Legros%2C%20V.%2C%20Chevreux%2C%20G.%2C%20Lacroix%2C%20B.%2C%20Castro%2C%20A.%2C%20Joly%2C%20N.%2C%20Lorca%2C%20T.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282025%29.%20The%20MAST%20kinase%20KIN-4%20carries%20out%20mitotic%20entry%20functions%20of%20Greatwall%20in%20C.%20elegans.%20%26lt%3Bi%26gt%3BThe%20EMBO%20Journal%26lt%3B%5C%2Fi%26gt%3B%2C%201%26%23×2013%3B32.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs44318-025-00364-w%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs44318-025-00364-w%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20MAST%20kinase%20KIN-4%20carries%20out%20mitotic%20entry%20functions%20of%20Greatwall%20in%20C.%20elegans%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ludivine%22%2C%22lastName%22%3A%22Roumbo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Batool%22%2C%22lastName%22%3A%22Ossareh-Nazari%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Suzanne%22%2C%22lastName%22%3A%22Vigneron%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ioanna%22%2C%22lastName%22%3A%22Stefani%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22V%5Cu00e9ronique%22%2C%22lastName%22%3A%22Legros%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Guillaume%22%2C%22lastName%22%3A%22Chevreux%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Benjamin%22%2C%22lastName%22%3A%22Lacroix%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anna%22%2C%22lastName%22%3A%22Castro%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thierry%22%2C%22lastName%22%3A%22Lorca%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22MAST-like%2C%20or%20Greatwall%20%28Gwl%29%2C%20an%20atypical%20protein%20kinase%20related%20to%20the%20evolutionarily%20conserved%20MAST%20kinase%20family%2C%20is%20crucial%20for%20cell%20cycle%20control%20during%20mitotic%20entry.%20Mechanistically%2C%20Greatwall%20is%20activated%20by%20Cyclin%20B-Cdk1%20phosphorylation%20of%20a%20550%20amino%20acids-long%20insertion%20in%20its%20atypical%20activation%20segment.%20Subsequently%2C%20Gwl%20phosphorylates%20Endosulfine%20and%20Arpp19%20to%20convert%20them%20into%20inhibitors%20of%20PP2A-B55%20phosphatase%2C%20thereby%20preventing%20early%20dephosphorylation%20of%20M-phase%20targets%20of%20Cyclin%20B-Cdk1.%20Here%2C%20searching%20for%20an%20elusive%20Gwl-like%20activity%20in%20C.%20elegans%2C%20we%20show%20that%20the%20single%20worm%20MAST%20kinase%2C%20KIN-4%2C%20fulfills%20this%20function%20in%20worms%20and%20can%20functionally%20replace%20Greatwall%20in%20the%20heterologous%20Xenopus%20system.%20Compared%20to%20Greatwall%2C%20the%20short%20activation%20segment%20of%20KIN-4%20lacks%20a%20phosphorylation%20site%2C%20and%20KIN-4%20is%20active%20even%20when%20produced%20in%20E.%20coli.%20We%20also%20show%20that%20a%20balance%20between%20Cyclin%20B-Cdk1%20and%20PP2A-B55%20activity%2C%20regulated%20by%20KIN-4%2C%20is%20essential%20to%20ensure%20asynchronous%20cell%20divisions%20in%20the%20early%20worm%20embryo.%20These%20findings%20resolve%20a%20long-standing%20puzzle%20related%20to%20the%20supposed%20absence%20of%20a%20Greatwall%20pathway%20in%20C.%20elegans%2C%20and%20highlight%20a%20novel%20aspect%20of%20PP2A-B55%20regulation%20by%20MAST%20kinases.%22%2C%22date%22%3A%222025-02-17%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs44318-025-00364-w%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.embopress.org%5C%2Fdoi%5C%2Ffull%5C%2F10.1038%5C%2Fs44318-025-00364-w%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220261-4189%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222025-02-18T09%3A31%3A22Z%22%7D%7D%2C%7B%22key%22%3A%22BQELGKWL%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22El%20Mossadeq%20et%20al.%22%2C%22parsedDate%22%3A%222024-12-26%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BEl%20Mossadeq%2C%20L.%2C%20Bellutti%2C%20L.%2C%20Le%20Borgne%2C%20R.%2C%20Canman%2C%20J.%20C.%2C%20Pintard%2C%20L.%2C%20Verbavatz%2C%20J.-M.%2C%20Askjaer%2C%20P.%2C%20%26amp%3B%20Dumont%2C%20J.%20%282024%29.%20An%20interkinetic%20envelope%20surrounds%20chromosomes%20between%20meiosis%20I%20and%20II%20in%20C.%20elegans%20oocytes.%20%26lt%3Bi%26gt%3BJournal%20of%20Cell%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B224%26lt%3B%5C%2Fi%26gt%3B%283%29%2C%20e202403125.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202403125%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202403125%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22An%20interkinetic%20envelope%20surrounds%20chromosomes%20between%20meiosis%20I%20and%20II%20in%20C.%20elegans%20oocytes%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Layla%22%2C%22lastName%22%3A%22El%20Mossadeq%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laura%22%2C%22lastName%22%3A%22Bellutti%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R%5Cu00e9mi%22%2C%22lastName%22%3A%22Le%20Borgne%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julie%20C.%22%2C%22lastName%22%3A%22Canman%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Marc%22%2C%22lastName%22%3A%22Verbavatz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%22%2C%22lastName%22%3A%22Askjaer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julien%22%2C%22lastName%22%3A%22Dumont%22%7D%5D%2C%22abstractNote%22%3A%22At%20the%20end%20of%20cell%20division%2C%20the%20nuclear%20envelope%20reassembles%20around%20the%20decondensing%20chromosomes.%20Female%20meiosis%20culminates%20in%20two%20consecutive%20cell%20divisions%20of%20the%20oocyte%2C%20meiosis%20I%20and%20II%2C%20which%20are%20separated%20by%20a%20brief%20transition%20phase%20known%20as%20interkinesis.%20Due%20to%20the%20absence%20of%20chromosome%20decondensation%20and%20the%20suppression%20of%20genome%20replication%20during%20interkinesis%2C%20it%20has%20been%20widely%20assumed%20that%20the%20nuclear%20envelope%20does%20not%20reassemble%20between%20meiosis%20I%20and%20II.%20By%20analyzing%20interkinesis%20in%20C.%20elegans%20oocytes%2C%20we%20instead%20show%20that%20an%20atypical%20structure%20made%20of%20two%20lipid%20bilayers%2C%20which%20we%20termed%20the%20interkinetic%20envelope%2C%20surrounds%20the%20surface%20of%20the%20segregating%20chromosomes.%20The%20interkinetic%20envelope%20shares%20common%20features%20with%20the%20nuclear%20envelope%20but%20also%20exhibits%20specific%20characteristics%20that%20distinguish%20it%2C%20including%20its%20lack%20of%20continuity%20with%20the%20endoplasmic%20reticulum%2C%20unique%20protein%20composition%2C%20assembly%20mechanism%2C%20and%20function%20in%20chromosome%20segregation.%20These%20distinct%20attributes%20collectively%20define%20the%20interkinetic%20envelope%20as%20a%20unique%20and%20specialized%20structure%20that%20has%20been%20previously%20overlooked.%22%2C%22date%22%3A%222024-12-26%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1083%5C%2Fjcb.202403125%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202403125%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220021-9525%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22L2N9KLHW%22%2C%22I7CUV6U5%22%2C%22MH4QXJHX%22%2C%22GWSKYQWE%22%2C%227DC6IIUA%22%5D%2C%22dateModified%22%3A%222025-01-02T10%3A15%3A31Z%22%7D%7D%2C%7B%22key%22%3A%22Z4276ISH%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Strzelecki%20et%20al.%22%2C%22parsedDate%22%3A%222024-09-28%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BStrzelecki%2C%20P.%2C%20Joly%2C%20N.%2C%20H%26%23xE9%3Bbraud%2C%20P.%2C%20Hoffmann%2C%20E.%2C%20Cech%2C%20G.%20M.%2C%20Kloska%2C%20A.%2C%20Busi%2C%20F.%2C%20%26amp%3B%20Grange%2C%20W.%20%282024%29.%20Enhanced%20Golden%20Gate%20Assembly%3A%20evaluating%20overhang%20strength%20for%20improved%20ligation%20efficiency.%20%26lt%3Bi%26gt%3BNucleic%20Acids%20Research%26lt%3B%5C%2Fi%26gt%3B%2C%20gkae809.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkae809%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkae809%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Enhanced%20Golden%20Gate%20Assembly%3A%20evaluating%20overhang%20strength%20for%20improved%20ligation%20efficiency%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Patryk%22%2C%22lastName%22%3A%22Strzelecki%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Pascal%22%2C%22lastName%22%3A%22H%5Cu00e9braud%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Elise%22%2C%22lastName%22%3A%22Hoffmann%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Grzegorz%5Cu00a0M%22%2C%22lastName%22%3A%22Cech%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anna%22%2C%22lastName%22%3A%22Kloska%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Florent%22%2C%22lastName%22%3A%22Busi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Wilfried%22%2C%22lastName%22%3A%22Grange%22%7D%5D%2C%22abstractNote%22%3A%22Molecular%20cloning%2C%20a%20routine%20yet%20essential%20technique%2C%20relies%20heavily%20on%20efficient%20ligation%2C%20which%20can%20be%20significantly%20improved%20using%20Golden%20Gate%5Cu00a0Assembly%20%28GGA%29.%20A%20key%20component%20of%20GGA%20is%20the%20use%20of%20type%20IIS%20enzymes%2C%20which%20uniquely%20cleave%20downstream%20of%20their%20recognition%20sequences%20to%20generate%20various%20overhangs%2C%20including%20non-palindromic%20ones.%20Recent%20advancements%20in%20GGA%20include%20the%20development%20of%20newly%20engineered%20enzymes%20with%20enhanced%20activity.%20Additionally%2C%20high-throughput%20GGA%20assays%2C%20which%20allow%20for%20the%20simultaneous%20study%20of%20all%20possible%20overhangs%2C%20have%20identified%20optimal%20GGA%20substrates%20with%20high%20efficiencies%20and%20fidelities%2C%20greatly%20facilitating%20the%20design%20of%20complex%20assemblies.%20Interestingly%2C%20these%20assays%20reveal%20unexpected%20correlations%20between%20ligation%20efficiencies%20and%20overhang%20stabilities.%20One%20hypothesis%20for%20this%20observation%20is%20that%20newly%20hydrolyzed%20DNA%20fragments%20with%20strong%20overhangs%20can%20readily%20re-ligate%2C%20thereby%20slowing%20down%20the%20overall%20process.%20In%20this%20paper%2C%20we%20employ%20a%20combination%20of%20gel%20electrophoresis%20and%20numerical%20calculations%20to%20test%20this%20hypothesis%2C%20ultimately%20determining%20that%20it%20does%20not%20hold%20true%20under%20the%20conditions%20established%20by%20conventional%20GGA%20assays.%20Using%20an%20assembly%20of%2010%20fragments%2C%20we%20demonstrate%20that%20strong%20overhangs%20yield%20higher%20GGA%20efficiency%2C%20while%20weak%20overhangs%20result%20in%20lower%20efficiency.%20These%20findings%20enable%20us%20to%20propose%20optimal%20overhangs%20for%20efficient%20GGA%20assays%2C%20significantly%20increasing%20yield.%22%2C%22date%22%3A%222024-09-28%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1093%5C%2Fnar%5C%2Fgkae809%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkae809%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220305-1048%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222024-09-30T08%3A55%3A27Z%22%7D%7D%2C%7B%22key%22%3A%229YPAUBFG%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Beaumale%20et%20al.%22%2C%22parsedDate%22%3A%222024-02-08%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BBeaumale%2C%20E.%2C%20Van%20Hove%2C%20L.%2C%20Pintard%2C%20L.%2C%20%26amp%3B%20Joly%2C%20N.%20%282024%29.%20Microtubule-binding%20domains%20in%20Katanin%20p80%20subunit%20are%20essential%20for%20severing%20activity%20in%20C.%20elegans.%20%26lt%3Bi%26gt%3BJournal%20of%20Cell%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B223%26lt%3B%5C%2Fi%26gt%3B%284%29%2C%20e202308023.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-ItemURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202308023%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202308023%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Microtubule-binding%20domains%20in%20Katanin%20p80%20subunit%20are%20essential%20for%20severing%20activity%20in%20C.%20elegans%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eva%22%2C%22lastName%22%3A%22Beaumale%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%5D%2C%22abstractNote%22%3A%22Microtubule-severing%20enzymes%20%28MSEs%29%2C%20such%20as%20Katanin%2C%20Spastin%2C%20and%20Fidgetin%20play%20essential%20roles%20in%20cell%20division%20and%20neurogenesis.%20They%20damage%20the%20microtubule%20%28MT%29%20lattice%2C%20which%20can%20either%20destroy%20or%20amplify%20the%20MT%20cytoskeleton%2C%20depending%20on%20the%20cellular%20context.%20However%2C%20little%20is%20known%20about%20how%20they%20interact%20with%20their%20substrates.%20We%20have%20identified%20the%20microtubule-binding%20domains%20%28MTBD%29%20required%20for%20Katanin%20function%20in%20C.%20elegans.%20Katanin%20is%20a%20heterohexamer%20of%20dimers%20containing%20a%20catalytic%20subunit%20p60%20and%20a%20regulatory%20subunit%20p80%2C%20both%20of%20which%20are%20essential%20for%20female%20meiotic%20spindle%20assembly.%20Here%2C%20we%20report%20that%20p80-like%28MEI-2%29%20dictates%20Katanin%20binding%20to%20MTs%20via%20two%20MTBDs%20composed%20of%20basic%20patches.%20Substituting%20these%20patches%20reduces%20Katanin%20binding%20to%20MTs%2C%20compromising%20its%20function%20in%20female%20meiotic-spindle%20assembly.%20Structural%20alignments%20of%20p80-like%28MEI-2%29%20with%20p80s%20from%20different%20species%20revealed%20that%20the%20MTBDs%20are%20evolutionarily%20conserved%2C%20even%20if%20the%20specific%20amino%20acids%20involved%20vary.%20Our%20findings%20highlight%20the%20critical%20importance%20of%20the%20regulatory%20subunit%20%28p80%29%20in%20providing%20MT%20binding%20to%20the%20Katanin%20complex.%22%2C%22date%22%3A%222024-02-08%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1083%5C%2Fjcb.202308023%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.202308023%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%220021-9525%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222024-02-13T08%3A46%3A43Z%22%7D%7D%2C%7B%22key%22%3A%22JMRE4R57%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Nkombo%20Nkoula%20et%20al.%22%2C%22parsedDate%22%3A%222023-07-19%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BNkombo%20Nkoula%2C%20S.%2C%20Velez-Aguilera%2C%20G.%2C%20Ossareh-Nazari%2C%20B.%2C%20Van%20Hove%2C%20L.%2C%20Ayuso%2C%20C.%2C%20Legros%2C%20V.%2C%20Chevreux%2C%20G.%2C%20Thomas%2C%20L.%2C%20Seydoux%2C%20G.%2C%20Askjaer%2C%20P.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282023%29.%20Mechanisms%20of%20nuclear%20pore%20complex%20disassembly%20by%20the%20mitotic%20Polo-like%20kinase%201%20%28PLK-1%29%20in%20C.%20elegans%20embryos.%20%26lt%3Bi%26gt%3BScience%20Advances%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B9%26lt%3B%5C%2Fi%26gt%3B%2829%29%2C%20eadf7826.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1126%5C%2Fsciadv.adf7826%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1126%5C%2Fsciadv.adf7826%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Mechanisms%20of%20nuclear%20pore%20complex%20disassembly%20by%20the%20mitotic%20Polo-like%20kinase%201%20%28PLK-1%29%20in%20C.%20elegans%20embryos%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sylvia%22%2C%22lastName%22%3A%22Nkombo%20Nkoula%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Griselda%22%2C%22lastName%22%3A%22Velez-Aguilera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Batool%22%2C%22lastName%22%3A%22Ossareh-Nazari%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cristina%22%2C%22lastName%22%3A%22Ayuso%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22V%5Cu00e9ronique%22%2C%22lastName%22%3A%22Legros%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Guillaume%22%2C%22lastName%22%3A%22Chevreux%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Laura%22%2C%22lastName%22%3A%22Thomas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22G%5Cu00e9raldine%22%2C%22lastName%22%3A%22Seydoux%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%22%2C%22lastName%22%3A%22Askjaer%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22The%20nuclear%20envelope%2C%20which%20protects%20and%20organizes%20the%20genome%2C%20is%20dismantled%20during%20mitosis.%20In%20the%20Caenorhabditis%20elegans%20zygote%2C%20nuclear%20envelope%20breakdown%20%28NEBD%29%20of%20the%20parental%20pronuclei%20is%20spatially%20and%20temporally%20regulated%20during%20mitosis%20to%20promote%20the%20unification%20of%20the%20maternal%20and%20paternal%20genomes.%20Nuclear%20pore%20complex%20%28NPC%29%20disassembly%20is%20a%20decisive%20step%20of%20NEBD%2C%20essential%20for%20nuclear%20permeabilization.%20By%20combining%20live%20imaging%2C%20biochemistry%2C%20and%20phosphoproteomics%2C%20we%20show%20that%20NPC%20disassembly%20is%20a%20stepwise%20process%20that%20involves%20Polo-like%20kinase%201%20%28PLK-1%29%5Cu2013dependent%20and%20%5Cu2013independent%20steps.%20PLK-1%20targets%20multiple%20NPC%20subcomplexes%2C%20including%20the%20cytoplasmic%20filaments%2C%20central%20channel%2C%20and%20inner%20ring.%20PLK-1%20is%20recruited%20to%20and%20phosphorylates%20intrinsically%20disordered%20regions%20%28IDRs%29%20of%20several%20multivalent%20linker%20nucleoporins.%20Notably%2C%20although%20the%20phosphosites%20are%20not%20conserved%20between%20human%20and%20C.%20elegans%20nucleoporins%2C%20they%20are%20located%20in%20IDRs%20in%20both%20species.%20Our%20results%20suggest%20that%20targeting%20IDRs%20of%20multivalent%20linker%20nucleoporins%20is%20an%20evolutionarily%20conserved%20driver%20of%20NPC%20disassembly%20during%20mitosis.%22%2C%22date%22%3A%222023-07-19%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1126%5C%2Fsciadv.adf7826%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fwww.science.org%5C%2Fdoi%5C%2F10.1126%5C%2Fsciadv.adf7826%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%22%22%2C%22language%22%3A%22%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222023-07-21T12%3A26%3A04Z%22%7D%7D%2C%7B%22key%22%3A%227SJ3YXZD%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Kouranti%20et%20al.%22%2C%22parsedDate%22%3A%222022-05-05%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BKouranti%2C%20I.%2C%20Abdel%20Khalek%2C%20W.%2C%20Mazurkiewicz%2C%20S.%2C%20Loisel-Ferreira%2C%20I.%2C%20Gautreau%2C%20A.%20M.%2C%20Pintard%2C%20L.%2C%20Jeunemaitre%2C%20X.%2C%20%26amp%3B%20Clauser%2C%20E.%20%282022%29.%20Cullin%203%20Exon%209%20Deletion%20in%20Familial%20Hyperkalemic%20Hypertension%20Impairs%20Cullin3-Ring-E3%20Ligase%20%28CRL3%29%20Dynamic%20Regulation%20and%20Cycling.%20%26lt%3Bi%26gt%3BInternational%20Journal%20of%20Molecular%20Sciences%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B23%26lt%3B%5C%2Fi%26gt%3B%289%29%2C%205151.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fijms23095151%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.3390%5C%2Fijms23095151%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Cullin%203%20Exon%209%20Deletion%20in%20Familial%20Hyperkalemic%20Hypertension%20Impairs%20Cullin3-Ring-E3%20Ligase%20%28CRL3%29%20Dynamic%20Regulation%20and%20Cycling%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ilektra%22%2C%22lastName%22%3A%22Kouranti%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Waed%22%2C%22lastName%22%3A%22Abdel%20Khalek%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Stephani%22%2C%22lastName%22%3A%22Mazurkiewicz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Irmine%22%2C%22lastName%22%3A%22Loisel-Ferreira%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Alexis%20M.%22%2C%22lastName%22%3A%22Gautreau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Xavier%22%2C%22lastName%22%3A%22Jeunemaitre%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eric%22%2C%22lastName%22%3A%22Clauser%22%7D%5D%2C%22abstractNote%22%3A%22Cullin%203%20%28CUL3%29%20is%20the%20scaffold%20of%20Cullin3%20Ring%20E3-ligases%20%28CRL3s%29%2C%20which%20use%20various%20BTB-adaptor%20proteins%20to%20ubiquitinate%20numerous%20substrates%20targeting%20their%20proteasomal%20degradation.%20CUL3%20mutations%2C%20responsible%20for%20a%20severe%20form%20of%20familial%20hyperkalemia%20and%20hypertension%20%28FHHt%29%2C%20all%20result%20in%20a%20deletion%20of%20exon%209%20%28amino-acids%20403-459%29%20%28CUL3-%5Cu22069%29.%20Surprisingly%2C%20while%20CUL3-%5Cu22069%20is%20hyperneddylated%2C%20a%20post-translational%20modification%20that%20typically%20activates%20CRL%20complexes%2C%20it%20is%20unable%20to%20ubiquitinate%20its%20substrates.%20In%20order%20to%20understand%20the%20mechanisms%20behind%20this%20loss-of%20function%2C%20we%20performed%20comparative%20label-free%20quantitative%20analyses%20of%20CUL3%20and%20CUL3-%5Cu22069%20interactome%20by%20mass%20spectrometry.%20It%20was%20observed%20that%20CUL3-%5Cu22069%20interactions%20with%20COP9%20and%20CAND1%2C%20both%20involved%20in%20CRL3%20complexes%26%23039%3B%20dynamic%20assembly%2C%20were%20disrupted.%20These%20defects%20result%20in%20a%20reduction%20in%20the%20dynamic%20cycling%20of%20the%20CRL3%20complexes%2C%20making%20the%20CRL3-%5Cu22069%20complex%20an%20inactive%20BTB-adaptor%20trap%2C%20as%20demonstrated%20by%20SILAC%20experiments.%20Collectively%2C%20the%20data%20indicated%20that%20the%20hyperneddylated%20CUL3-%5Cu22069%20protein%20is%20inactive%20as%20a%20consequence%20of%20several%20structural%20changes%20disrupting%20its%20dynamic%20interactions%20with%20key%20regulatory%20partners.%22%2C%22date%22%3A%222022-05-05%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.3390%5C%2Fijms23095151%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221422-0067%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A25%3A09Z%22%7D%7D%2C%7B%22key%22%3A%22GI4PXUMF%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Velez-Aguilera%20et%20al.%22%2C%22parsedDate%22%3A%222022-03-08%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVelez-Aguilera%2C%20G.%2C%20Ossareh-Nazari%2C%20B.%2C%20Van%20Hove%2C%20L.%2C%20Joly%2C%20N.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282022%29.%20Cortical%20microtubule%20pulling%20forces%20contribute%20to%20the%20union%20of%20the%20parental%20genomes%20in%20the%20Caenorhabditis%20elegans%20zygote.%20%26lt%3Bi%26gt%3BeLife%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B11%26lt%3B%5C%2Fi%26gt%3B%2C%20e75382.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.75382%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.75382%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Cortical%20microtubule%20pulling%20forces%20contribute%20to%20the%20union%20of%20the%20parental%20genomes%20in%20the%20Caenorhabditis%20elegans%20zygote%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Griselda%22%2C%22lastName%22%3A%22Velez-Aguilera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Batool%22%2C%22lastName%22%3A%22Ossareh-Nazari%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22Previously%2C%20we%20reported%20that%20the%20Polo-like%20kinase%20PLK-1%20phosphorylates%20the%20single%20Caenorhabditis%20elegans%20lamin%20%28LMN-1%29%20to%20trigger%20lamina%20depolymerization%20during%20mitosis.%20We%20showed%20that%20this%20event%20is%20required%20to%20form%20a%20pronuclear%20envelope%20scission%20event%20that%20removes%20membranes%20on%20the%20juxtaposed%20oocyte%20and%20sperm%20pronuclear%20envelopes%20in%20the%20zygote%2C%20allowing%20the%20parental%20chromosomes%20to%20merge%20in%20a%20single%20nucleus%20after%20segregation%20%28Velez-Aguilera%20et%20al.%2C%202020%29.%20Here%2C%20we%20show%20that%20cortical%20microtubule%20pulling%20forces%20contribute%20to%20pronuclear%20envelopes%20scission%20by%20promoting%20mitotic%20spindle%20elongation%2C%20and%20conversely%2C%20nuclear%20envelopes%20remodeling%20facilitates%20spindle%20elongation.%20We%20also%20demonstrate%20that%20weakening%20the%20pronuclear%20envelopes%20via%20PLK-1-mediated%20lamina%20depolymerization%2C%20is%20a%20prerequisite%20for%20the%20astral%20microtubule%20pulling%20forces%20to%20trigger%20pronuclear%20membranes%20scission.%20Finally%2C%20we%20provide%20evidence%20that%20PLK-1%20mainly%20acts%20via%20lamina%20depolymerization%20in%20this%20process.%20These%20observations%20thus%20indicate%20that%20temporal%20coordination%20between%20lamina%20depolymerization%20and%20mitotic%20spindle%20elongation%20facilitates%20pronuclear%20envelopes%20scission%20and%20parental%20genomes%20unification.%22%2C%22date%22%3A%222022-03-08%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.7554%5C%2FeLife.75382%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222050-084X%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A09Z%22%7D%7D%2C%7B%22key%22%3A%22R2DYQDHX%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Knox%20et%20al.%22%2C%22parsedDate%22%3A%222021-04-28%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BKnox%2C%20J.%2C%20Joly%2C%20N.%2C%20Linossi%2C%20E.%20M.%2C%20Carmona-Negr%26%23xF3%3Bn%2C%20J.%20A.%2C%20Jura%2C%20N.%2C%20Pintard%2C%20L.%2C%20Zuercher%2C%20W.%2C%20%26amp%3B%20Roy%2C%20P.%20J.%20%282021%29.%20A%20survey%20of%20the%20kinome%20pharmacopeia%20reveals%20multiple%20scaffolds%20and%20targets%20for%20the%20development%20of%20novel%20anthelmintics.%20%26lt%3Bi%26gt%3BScientific%20Reports%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B11%26lt%3B%5C%2Fi%26gt%3B%281%29%2C%209161.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41598-021-88150-6%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41598-021-88150-6%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20survey%20of%20the%20kinome%20pharmacopeia%20reveals%20multiple%20scaffolds%20and%20targets%20for%20the%20development%20of%20novel%20anthelmintics%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jessica%22%2C%22lastName%22%3A%22Knox%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Edmond%20M.%22%2C%22lastName%22%3A%22Linossi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jos%5Cu00e9%20A.%22%2C%22lastName%22%3A%22Carmona-Negr%5Cu00f3n%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Natalia%22%2C%22lastName%22%3A%22Jura%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22William%22%2C%22lastName%22%3A%22Zuercher%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Peter%20J.%22%2C%22lastName%22%3A%22Roy%22%7D%5D%2C%22abstractNote%22%3A%22Over%20one%20billion%20people%20are%20currently%20infected%20with%20a%20parasitic%20nematode.%20Symptoms%20can%20include%20anemia%2C%20malnutrition%2C%20developmental%20delay%2C%20and%20in%20severe%20cases%2C%20death.%20Resistance%20is%20emerging%20to%20the%20anthelmintics%20currently%20used%20to%20treat%20nematode%20infection%2C%20prompting%20the%20need%20to%20develop%20new%20anthelmintics.%20Towards%20this%20end%2C%20we%20identified%20a%20set%20of%20kinases%20that%20may%20be%20targeted%20in%20a%20nematode-selective%20manner.%20We%20first%20screened%202040%20inhibitors%20of%20vertebrate%20kinases%20for%20those%20that%20impair%20the%20model%20nematode%20Caenorhabditis%20elegans.%20By%20determining%20whether%20the%20terminal%20phenotype%20induced%20by%20each%20kinase%20inhibitor%20matched%20that%20of%20the%20predicted%20target%20mutant%20in%20C.%20elegans%2C%20we%20identified%2017%20druggable%20nematode%20kinase%20targets.%20Of%20these%2C%20we%20found%20that%20nematode%20EGFR%2C%20MEK1%2C%20and%20PLK1%20kinases%20have%20diverged%20from%20vertebrates%20within%20their%20drug-binding%20pocket.%20For%20each%20of%20these%20targets%2C%20we%20identified%20small%20molecule%20scaffolds%20that%20may%20be%20further%20modified%20to%20develop%20nematode-selective%20inhibitors.%20Nematode%20EGFR%2C%20MEK1%2C%20and%20PLK1%20therefore%20represent%20key%20targets%20for%20the%20development%20of%20new%20anthelmintic%20medicines.%22%2C%22date%22%3A%222021-04-28%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41598-021-88150-6%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222045-2322%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A24Z%22%7D%7D%2C%7B%22key%22%3A%2287GI54GG%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Tavernier%20et%20al.%22%2C%22parsedDate%22%3A%222021-03-26%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BTavernier%2C%20N.%2C%20Thomas%2C%20Y.%2C%20Vigneron%2C%20S.%2C%20Maisonneuve%2C%20P.%2C%20Orlicky%2C%20S.%2C%20Mader%2C%20P.%2C%20Regmi%2C%20S.%20G.%2C%20Van%20Hove%2C%20L.%2C%20Levinson%2C%20N.%20M.%2C%20Gasmi-Seabrook%2C%20G.%2C%20Joly%2C%20N.%2C%20Poteau%2C%20M.%2C%20Velez-Aguilera%2C%20G.%2C%20Gavet%2C%20O.%2C%20Castro%2C%20A.%2C%20Dasso%2C%20M.%2C%20Lorca%2C%20T.%2C%20Sicheri%2C%20F.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282021%29.%20Bora%20phosphorylation%20substitutes%20in%20trans%20for%20T-loop%20phosphorylation%20in%20Aurora%20A%20to%20promote%20mitotic%20entry.%20%26lt%3Bi%26gt%3BNature%20Communications%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B12%26lt%3B%5C%2Fi%26gt%3B%281%29%2C%201899.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41467-021-21922-w%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1038%5C%2Fs41467-021-21922-w%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Bora%20phosphorylation%20substitutes%20in%20trans%20for%20T-loop%20phosphorylation%20in%20Aurora%20A%20to%20promote%20mitotic%20entry%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22N.%22%2C%22lastName%22%3A%22Tavernier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Y.%22%2C%22lastName%22%3A%22Thomas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%22%2C%22lastName%22%3A%22Vigneron%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22P.%22%2C%22lastName%22%3A%22Maisonneuve%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%22%2C%22lastName%22%3A%22Orlicky%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22P.%22%2C%22lastName%22%3A%22Mader%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S.%20G.%22%2C%22lastName%22%3A%22Regmi%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L.%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22N.%20M.%22%2C%22lastName%22%3A%22Levinson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22G.%22%2C%22lastName%22%3A%22Gasmi-Seabrook%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22N.%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M.%22%2C%22lastName%22%3A%22Poteau%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22G.%22%2C%22lastName%22%3A%22Velez-Aguilera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22O.%22%2C%22lastName%22%3A%22Gavet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22A.%22%2C%22lastName%22%3A%22Castro%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M.%22%2C%22lastName%22%3A%22Dasso%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22T.%22%2C%22lastName%22%3A%22Lorca%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22F.%22%2C%22lastName%22%3A%22Sicheri%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22L.%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22Polo-like%20kinase%201%20%28Plk1%29%20is%20instrumental%20for%20mitotic%20entry%20and%20progression.%20Plk1%20is%20activated%20by%20phosphorylation%20on%20a%20conserved%20residue%20Thr210%20in%20its%20activation%20segment%20by%20the%20Aurora%20A%20kinase%20%28AURKA%29%2C%20a%20reaction%20that%20critically%20requires%20the%20co-factor%20Bora%20phosphorylated%20by%20a%20CyclinA%5C%2FB-Cdk1%20kinase.%20Here%20we%20show%20that%20phospho-Bora%20is%20a%20direct%20activator%20of%20AURKA%20kinase%20activity.%20We%20localize%20the%20key%20determinants%20of%20phospho-Bora%20function%20to%20a%20100%20amino%20acid%20region%20encompassing%20two%20short%20Tpx2-like%20motifs%20and%20a%20phosphoSerine-Proline%20motif%20at%20Serine%20112%2C%20through%20which%20Bora%20binds%20AURKA.%20The%20latter%20substitutes%20in%20trans%20for%20the%20Thr288%20phospho-regulatory%20site%20of%20AURKA%2C%20which%20is%20essential%20for%20an%20active%20conformation%20of%20the%20kinase%20domain.%20We%20demonstrate%20the%20importance%20of%20these%20determinants%20for%20Bora%20function%20in%20mitotic%20entry%20both%20in%20Xenopus%20egg%20extracts%20and%20in%5Cu00a0human%20cells.%20Our%20findings%20unveil%20the%20activation%20mechanism%20of%20AURKA%20that%20is%20critical%20for%20mitotic%20entry.%22%2C%22date%22%3A%222021-03-26%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1038%5C%2Fs41467-021-21922-w%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222041-1723%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A24Z%22%7D%7D%2C%7B%22key%22%3A%22JDPEX3E3%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Velez-Aguilera%20et%20al.%22%2C%22parsedDate%22%3A%222020-10-08%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVelez-Aguilera%2C%20G.%2C%20Nkombo%20Nkoula%2C%20S.%2C%20Ossareh-Nazari%2C%20B.%2C%20Link%2C%20J.%2C%20Paouneskou%2C%20D.%2C%20Van%20Hove%2C%20L.%2C%20Joly%2C%20N.%2C%20Tavernier%2C%20N.%2C%20Verbavatz%2C%20J.-M.%2C%20Jantsch%2C%20V.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282020%29.%20PLK-1%20promotes%20the%20merger%20of%20the%20parental%20genome%20into%20a%20single%20nucleus%20by%20triggering%20lamina%20disassembly.%20%26lt%3Bi%26gt%3BeLife%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B9%26lt%3B%5C%2Fi%26gt%3B%2C%20e59510.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.59510%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.7554%5C%2FeLife.59510%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22PLK-1%20promotes%20the%20merger%20of%20the%20parental%20genome%20into%20a%20single%20nucleus%20by%20triggering%20lamina%20disassembly%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Griselda%22%2C%22lastName%22%3A%22Velez-Aguilera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sylvia%22%2C%22lastName%22%3A%22Nkombo%20Nkoula%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Batool%22%2C%22lastName%22%3A%22Ossareh-Nazari%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jana%22%2C%22lastName%22%3A%22Link%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Dimitra%22%2C%22lastName%22%3A%22Paouneskou%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Tavernier%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Marc%22%2C%22lastName%22%3A%22Verbavatz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Verena%22%2C%22lastName%22%3A%22Jantsch%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22Life%20of%20sexually%20reproducing%20organisms%20starts%20with%20the%20fusion%20of%20the%20haploid%20egg%20and%20sperm%20gametes%20to%20form%20the%20genome%20of%20a%20new%20diploid%20organism.%20Using%20the%20newly%20fertilized%20Caenorhabditis%20elegans%20zygote%2C%20we%20show%20that%20the%20mitotic%20Polo-like%20kinase%20PLK-1%20phosphorylates%20the%20lamin%20LMN-1%20to%20promote%20timely%20lamina%20disassembly%20and%20subsequent%20merging%20of%20the%20parental%20genomes%20into%20a%20single%20nucleus%20after%20mitosis.%20Expression%20of%20non-phosphorylatable%20versions%20of%20LMN-1%2C%20which%20affect%20lamina%20depolymerization%20during%20mitosis%2C%20is%20sufficient%20to%20prevent%20the%20mixing%20of%20the%20parental%20chromosomes%20into%20a%20single%20nucleus%20in%20daughter%20cells.%20Finally%2C%20we%20recapitulate%20lamina%20depolymerization%20by%20PLK-1%20in%20vitro%20demonstrating%20that%20LMN-1%20is%20a%20direct%20PLK-1%20target.%20Our%20findings%20indicate%20that%20the%20timely%20removal%20of%20lamin%20is%20essential%20for%20the%20merging%20of%20parental%20chromosomes%20at%20the%20beginning%20of%20life%20in%20C.%20elegans%20and%20possibly%20also%20in%20humans%2C%20where%20a%20defect%20in%20this%20process%20might%20be%20fatal%20for%20embryo%20development.%22%2C%22date%22%3A%222020-10-08%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.7554%5C%2FeLife.59510%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222050-084X%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A39Z%22%7D%7D%2C%7B%22key%22%3A%229J76DXBR%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Joly%20et%20al.%22%2C%22parsedDate%22%3A%222020-06-01%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BJoly%2C%20N.%2C%20Beaumale%2C%20E.%2C%20Van%20Hove%2C%20L.%2C%20Martino%2C%20L.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282020%29.%20Phosphorylation%20of%20the%20microtubule-severing%20AAA%2B%20enzyme%20Katanin%20regulates%20C.%20elegans%20embryo%20development.%20%26lt%3Bi%26gt%3BThe%20Journal%20of%20Cell%20Biology%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B219%26lt%3B%5C%2Fi%26gt%3B%286%29%2C%20e201912037.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.201912037%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1083%5C%2Fjcb.201912037%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Phosphorylation%20of%20the%20microtubule-severing%20AAA%2B%20enzyme%20Katanin%20regulates%20C.%20elegans%20embryo%20development%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Eva%22%2C%22lastName%22%3A%22Beaumale%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lisa%22%2C%22lastName%22%3A%22Martino%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22The%20evolutionarily%20conserved%20microtubule%20%28MT%29-severing%20AAA-ATPase%20enzyme%20Katanin%20is%20emerging%20as%20a%20critical%20regulator%20of%20MT%20dynamics.%20In%20Caenorhabditis%20elegans%2C%20Katanin%20MT-severing%20activity%20is%20essential%20for%20meiotic%20spindle%20assembly%20but%20is%20toxic%20for%20the%20mitotic%20spindle.%20Here%20we%20analyzed%20Katanin%20dynamics%20in%20C.%20elegans%20and%20deciphered%20the%20role%20of%20Katanin%20phosphorylation%20in%20the%20regulation%20of%20its%20activity%20and%20stability.%20Katanin%20is%20abundant%20in%20oocytes%2C%20and%20its%20levels%20drop%20after%20meiosis%2C%20but%20unexpectedly%2C%20a%20significant%20fraction%20is%20present%20throughout%20embryogenesis%2C%20where%20it%20is%20dynamically%20recruited%20to%20the%20centrosomes%20and%20chromosomes%20during%20mitosis.%20We%20show%20that%20the%20minibrain%20kinase%20MBK-2%2C%20which%20is%20activated%20during%20meiosis%2C%20phosphorylates%20Katanin%20at%20multiple%20serines.%20We%20demonstrate%20unequivocally%20that%20Katanin%20phosphorylation%20at%20a%20single%20residue%20is%20necessary%20and%20sufficient%20to%20target%20Katanin%20for%20proteasomal%20degradation%20after%20meiosis%2C%20whereas%20phosphorylation%20at%20the%20other%20sites%20only%20inhibits%20Katanin%20ATPase%20activity%20stimulated%20by%20MTs.%20Our%20findings%20suggest%20that%20cycles%20of%20phosphorylation%20and%20dephosphorylation%20fine-tune%20Katanin%20level%20and%20activity%20to%20deliver%20the%20appropriate%20MT-severing%20activity%20during%20development.%22%2C%22date%22%3A%222020-06-01%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1083%5C%2Fjcb.201912037%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221540-8140%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A26%3A54Z%22%7D%7D%2C%7B%22key%22%3A%22IKTDRN4D%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Gutnik%20et%20al.%22%2C%22parsedDate%22%3A%222018-07-16%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BGutnik%2C%20S.%2C%20Thomas%2C%20Y.%2C%20Guo%2C%20Y.%2C%20Stoecklin%2C%20J.%2C%20Neagu%2C%20A.%2C%20Pintard%2C%20L.%2C%20Merlet%2C%20J.%2C%20%26amp%3B%20Ciosk%2C%20R.%20%282018%29.%20PRP-19%2C%20a%20conserved%20pre-mRNA%20processing%20factor%20and%20E3%20ubiquitin%20ligase%2C%20inhibits%20the%20nuclear%20accumulation%20of%20GLP-1%5C%2FNotch%20intracellular%20domain.%20%26lt%3Bi%26gt%3BBiology%20Open%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B7%26lt%3B%5C%2Fi%26gt%3B%287%29%2C%20bio034066.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1242%5C%2Fbio.034066%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1242%5C%2Fbio.034066%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22PRP-19%2C%20a%20conserved%20pre-mRNA%20processing%20factor%20and%20E3%20ubiquitin%20ligase%2C%20inhibits%20the%20nuclear%20accumulation%20of%20GLP-1%5C%2FNotch%20intracellular%20domain%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Silvia%22%2C%22lastName%22%3A%22Gutnik%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yann%22%2C%22lastName%22%3A%22Thomas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yanwu%22%2C%22lastName%22%3A%22Guo%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Janosch%22%2C%22lastName%22%3A%22Stoecklin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anca%22%2C%22lastName%22%3A%22Neagu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jorge%22%2C%22lastName%22%3A%22Merlet%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Rafal%22%2C%22lastName%22%3A%22Ciosk%22%7D%5D%2C%22abstractNote%22%3A%22The%20Notch%20signalling%20pathway%20is%20a%20conserved%20and%20widespread%20signalling%20paradigm%2C%20and%20its%20misregulation%20has%20been%20implicated%20in%20numerous%20disorders%2C%20including%20cancer.%20The%20output%20of%20Notch%20signalling%20depends%20on%20the%20nuclear%20accumulation%20of%20the%20Notch%20receptor%20intracellular%20domain%20%28ICD%29.%20Using%20the%20Caenorhabditis%20elegans%20germline%2C%20where%20GLP-1%5C%2FNotch-mediated%20signalling%20is%20essential%20for%20maintaining%20stem%20cells%2C%20we%20monitored%20GLP-1%20in%20vivo%20We%20found%20that%20the%20nuclear%20enrichment%20of%20GLP-1%20ICD%20is%20dynamic%3A%20while%20the%20ICD%20is%20enriched%20in%20germ%20cell%20nuclei%20during%20larval%20development%2C%20it%20is%20depleted%20from%20the%20nuclei%20in%20adult%20germlines.%20We%20found%20that%20this%20pattern%20depends%20on%20the%20ubiquitin%20proteolytic%20system%20and%20the%20splicing%20machinery%20and%2C%20identified%20the%20splicing%20factor%20PRP-19%20as%20a%20candidate%20E3%20ubiquitin%20ligase%20required%20for%20the%20nuclear%20depletion%20of%20GLP-1%20ICD.%22%2C%22date%22%3A%222018-07-16%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1242%5C%2Fbio.034066%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%222046-6390%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A35Z%22%7D%7D%2C%7B%22key%22%3A%227UENQK9Z%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Vigneron%20et%20al.%22%2C%22parsedDate%22%3A%222018-06-04%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BVigneron%2C%20S.%2C%20Sundermann%2C%20L.%2C%20Labb%26%23xE9%3B%2C%20J.-C.%2C%20Pintard%2C%20L.%2C%20Radulescu%2C%20O.%2C%20Castro%2C%20A.%2C%20%26amp%3B%20Lorca%2C%20T.%20%282018%29.%20Cyclin%20A-cdk1-Dependent%20Phosphorylation%20of%20Bora%20Is%20the%20Triggering%20Factor%20Promoting%20Mitotic%20Entry.%20%26lt%3Bi%26gt%3BDevelopmental%20Cell%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B45%26lt%3B%5C%2Fi%26gt%3B%285%29%2C%20637-650.e7.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2018.05.005%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2018.05.005%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Cyclin%20A-cdk1-Dependent%20Phosphorylation%20of%20Bora%20Is%20the%20Triggering%20Factor%20Promoting%20Mitotic%20Entry%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Suzanne%22%2C%22lastName%22%3A%22Vigneron%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lena%22%2C%22lastName%22%3A%22Sundermann%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Claude%22%2C%22lastName%22%3A%22Labb%5Cu00e9%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ovidiu%22%2C%22lastName%22%3A%22Radulescu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Anna%22%2C%22lastName%22%3A%22Castro%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thierry%22%2C%22lastName%22%3A%22Lorca%22%7D%5D%2C%22abstractNote%22%3A%22Mitosis%20is%20induced%20by%20the%20activation%20of%20the%20cyclin%20B%5C%2Fcdk1%20feedback%20loop%20that%20creates%20a%20bistable%20state.%20The%20triggering%20factor%20promoting%20active%20cyclin%20B%5C%2Fcdk1%20switch%20has%20been%20assigned%20to%20cyclin%20B%5C%2Fcdk1%20accumulation%20during%20G2.%20However%2C%20this%20complex%20is%20rapidly%20inactivated%20by%20Wee1%5C%2FMyt1-dependent%20phosphorylation%20of%20cdk1%20making%20unlikely%20a%20triggering%20role%20of%20this%20kinase%20in%20mitotic%20commitment.%20Here%20we%20show%20that%20cyclin%20A%5C%2Fcdk1%20kinase%20is%20the%20factor%20triggering%20mitosis.%20Cyclin%20A%5C%2Fcdk1%20phosphorylates%20Bora%20to%20promote%20Aurora%20A-dependent%20Plk1%20phosphorylation%20and%20activation%20and%20mitotic%20entry.%20We%20demonstrate%20that%20Bora%20phosphorylation%20by%20cyclin%20A%5C%2Fcdk1%20is%20both%20necessary%20and%20sufficient%20for%20mitotic%20commitment.%20Finally%2C%20we%20identify%20a%20site%20in%20Bora%20whose%20phosphorylation%20by%20cyclin%20A%5C%2Fcdk1%20is%20required%20for%20mitotic%20entry.%20We%20constructed%20a%20mathematical%20model%20confirming%20the%20essential%20role%20of%20this%20kinase%20in%20mitotic%20commitment.%20Overall%2C%20our%20results%20uncover%20the%20molecular%20mechanism%20by%20which%20cyclin%20A%5C%2Fcdk1%20triggers%20mitotic%20entry.%22%2C%22date%22%3A%222018-06-04%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.devcel.2018.05.005%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221878-1551%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A27%3A49Z%22%7D%7D%2C%7B%22key%22%3A%226KAAASYL%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Martino%20et%20al.%22%2C%22parsedDate%22%3A%222017-10-23%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BMartino%2C%20L.%2C%20Morchoisne-Bolhy%2C%20S.%2C%20Cheerambathur%2C%20D.%20K.%2C%20Van%20Hove%2C%20L.%2C%20Dumont%2C%20J.%2C%20Joly%2C%20N.%2C%20Desai%2C%20A.%2C%20Doye%2C%20V.%2C%20%26amp%3B%20Pintard%2C%20L.%20%282017%29.%20Channel%20Nucleoporins%20Recruit%20PLK-1%20to%20Nuclear%20Pore%20Complexes%20to%20Direct%20Nuclear%20Envelope%20Breakdown%20in%20C.%26%23xA0%3Belegans.%20%26lt%3Bi%26gt%3BDevelopmental%20Cell%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B43%26lt%3B%5C%2Fi%26gt%3B%282%29%2C%20157-171.e7.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2017.09.019%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2017.09.019%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Channel%20Nucleoporins%20Recruit%20PLK-1%20to%20Nuclear%20Pore%20Complexes%20to%20Direct%20Nuclear%20Envelope%20Breakdown%20in%20C.%5Cu00a0elegans%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lisa%22%2C%22lastName%22%3A%22Martino%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22St%5Cu00e9phanie%22%2C%22lastName%22%3A%22Morchoisne-Bolhy%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Dhanya%20K.%22%2C%22lastName%22%3A%22Cheerambathur%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lucie%22%2C%22lastName%22%3A%22Van%20Hove%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julien%22%2C%22lastName%22%3A%22Dumont%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Arshad%22%2C%22lastName%22%3A%22Desai%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Val%5Cu00e9rie%22%2C%22lastName%22%3A%22Doye%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%5D%2C%22abstractNote%22%3A%22In%20animal%20cells%2C%20nuclear%20envelope%20breakdown%20%28NEBD%29%20is%20required%20for%20proper%20chromosome%20segregation.%20Whereas%20mitotic%20kinases%20have%20been%20implicated%20in%20NEBD%2C%20how%20they%20coordinate%20their%20activity%20to%20trigger%20this%20event%20is%20unclear.%20Here%2C%20we%20show%20that%20both%20in%20human%20cells%20and%20Caenorhabditis%20elegans%2C%20the%20Polo-like%20kinase%201%20%28PLK-1%29%20is%20recruited%20to%20the%20nuclear%20pore%20complexes%2C%20just%20prior%20to%20NEBD%2C%20through%20its%20Polo-box%20domain%20%28PBD%29.%20We%20provide%20evidence%20that%20PLK-1%20localization%20to%20the%20nuclear%20envelope%20%28NE%29%20is%20required%20for%20efficient%20NEBD.%20We%20identify%20the%20central%20channel%5Cu00a0nucleoporins%20NPP-1%5C%2FNup58%2C%20NPP-4%5C%2FNup54%2C%20and%20NPP-11%5C%2FNup62%20as%20the%20critical%20factors%20anchoring%20PLK-1%20to%20the%20NE%20in%20C.%5Cu00a0elegans.%20In%20particular%2C%20NPP-1%2C%20NPP-4%2C%20and%20NPP-11%20primed%20at%20multiple%20Polo-docking%20sites%20by%20Cdk1%20and%20PLK-1%20itself%20physically%20interact%20with%20the%20PLK-1%20PBD.%20We%20conclude%20that%20nucleoporins%20play%20an%20unanticipated%20regulatory%20role%20in%20NEBD%2C%20by%20recruiting%20PLK-1%20to%20the%20NE%20thereby%20facilitating%20phosphorylation%20of%20critical%20downstream%20targets.%22%2C%22date%22%3A%222017-10-23%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.devcel.2017.09.019%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221878-1551%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222023-05-02T12%3A21%3A57Z%22%7D%7D%2C%7B%22key%22%3A%22RWWWZVZ8%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Dickinson%20et%20al.%22%2C%22parsedDate%22%3A%222017-08-21%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BDickinson%2C%20D.%20J.%2C%20Schwager%2C%20F.%2C%20Pintard%2C%20L.%2C%20Gotta%2C%20M.%2C%20%26amp%3B%20Goldstein%2C%20B.%20%282017%29.%20A%20Single-Cell%20Biochemistry%20Approach%20Reveals%20PAR%20Complex%20Dynamics%20during%20Cell%20Polarization.%20%26lt%3Bi%26gt%3BDevelopmental%20Cell%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B42%26lt%3B%5C%2Fi%26gt%3B%284%29%2C%20416-434.e11.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2017.07.024%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.devcel.2017.07.024%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22A%20Single-Cell%20Biochemistry%20Approach%20Reveals%20PAR%20Complex%20Dynamics%20during%20Cell%20Polarization%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Daniel%20J.%22%2C%22lastName%22%3A%22Dickinson%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Francoise%22%2C%22lastName%22%3A%22Schwager%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Lionel%22%2C%22lastName%22%3A%22Pintard%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Monica%22%2C%22lastName%22%3A%22Gotta%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bob%22%2C%22lastName%22%3A%22Goldstein%22%7D%5D%2C%22abstractNote%22%3A%22Regulated%20protein-protein%20interactions%20are%20critical%20for%20cell%20signaling%2C%20differentiation%2C%20and%20development.%20For%20the%20study%20of%20dynamic%20regulation%20of%20protein%20interactions%20in%5Cu00a0vivo%2C%20there%20is%20a%20need%20for%20techniques%20that%20can%20yield%20time-resolved%20information%20and%20probe%20multiple%20protein%20binding%20partners%20simultaneously%2C%20using%20small%20amounts%20of%20starting%20material.%20Here%20we%20describe%20a%20single-cell%20protein%20interaction%20assay.%20Single-cell%20lysates%20are%20generated%20at%20defined%20time%20points%20and%20analyzed%20using%20single-molecule%20pull-down%2C%20yielding%20information%20about%20dynamic%20protein%20complex%20regulation%20in%5Cu00a0vivo.%20We%20established%20the%20utility%20of%20this%20approach%20by%20studying%20PAR%20polarity%20proteins%2C%20which%20mediate%20polarization%20of%20many%20animal%20cell%20types.%20We%20uncovered%20striking%20regulation%20of%20PAR%20complex%20composition%20and%20stoichiometry%20during%20Caenorhabditis%20elegans%20zygote%20polarization%2C%20which%20takes%20place%20in%20less%20than%2020%5Cu00a0min.%20PAR%20complex%20dynamics%20are%20linked%20to%20the%20cell%20cycle%20by%20Polo-like%20kinase%201%20and%20govern%20the%20movement%20of%20PAR%20proteins%20to%20establish%20polarity.%20Our%20results%20demonstrate%20an%20approach%20to%20study%20dynamic%20biochemical%20events%20in%5Cu00a0vivo.%22%2C%22date%22%3A%222017-08-21%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.devcel.2017.07.024%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221878-1551%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A28%3A03Z%22%7D%7D%2C%7B%22key%22%3A%22CNKI2D58%22%2C%22library%22%3A%7B%22id%22%3A2913254%7D%2C%22meta%22%3A%7B%22lastModifiedByUser%22%3A%7B%22id%22%3A11274337%2C%22username%22%3A%22Charlotte_Brancaz%22%2C%22name%22%3A%22%22%2C%22links%22%3A%7B%22alternate%22%3A%7B%22href%22%3A%22https%3A%5C%2F%5C%2Fwww.zotero.org%5C%2Fcharlotte_brancaz%22%2C%22type%22%3A%22text%5C%2Fhtml%22%7D%7D%7D%2C%22creatorSummary%22%3A%22Richarme%20et%20al.%22%2C%22parsedDate%22%3A%222017-07-14%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%26lt%3Bdiv%20class%3D%26quot%3Bcsl-bib-body%26quot%3B%20style%3D%26quot%3Bline-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%26quot%3B%26gt%3B%5Cn%20%20%26lt%3Bdiv%20class%3D%26quot%3Bcsl-entry%26quot%3B%26gt%3BRicharme%2C%20G.%2C%20Liu%2C%20C.%2C%20Mihoub%2C%20M.%2C%20Abdallah%2C%20J.%2C%20Leger%2C%20T.%2C%20Joly%2C%20N.%2C%20Liebart%2C%20J.-C.%2C%20Jurkunas%2C%20U.%20V.%2C%20Nadal%2C%20M.%2C%20Bouloc%2C%20P.%2C%20Dairou%2C%20J.%2C%20%26amp%3B%20Lamouri%2C%20A.%20%282017%29.%20Guanine%20glycation%20repair%20by%20DJ-1%5C%2FPark7%20and%20its%20bacterial%20homologs.%20%26lt%3Bi%26gt%3BScience%20%28New%20York%2C%20N.Y.%29%26lt%3B%5C%2Fi%26gt%3B%2C%20%26lt%3Bi%26gt%3B357%26lt%3B%5C%2Fi%26gt%3B%286347%29%2C%20208%26%23×2013%3B211.%20%26lt%3Ba%20class%3D%26%23039%3Bzp-DOIURL%26%23039%3B%20href%3D%26%23039%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1126%5C%2Fscience.aag1095%26%23039%3B%26gt%3Bhttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1126%5C%2Fscience.aag1095%26lt%3B%5C%2Fa%26gt%3B%26lt%3B%5C%2Fdiv%26gt%3B%5Cn%26lt%3B%5C%2Fdiv%26gt%3B%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Guanine%20glycation%20repair%20by%20DJ-1%5C%2FPark7%20and%20its%20bacterial%20homologs%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Gilbert%22%2C%22lastName%22%3A%22Richarme%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Cailing%22%2C%22lastName%22%3A%22Liu%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mouadh%22%2C%22lastName%22%3A%22Mihoub%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jad%22%2C%22lastName%22%3A%22Abdallah%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Thibaut%22%2C%22lastName%22%3A%22Leger%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Nicolas%22%2C%22lastName%22%3A%22Joly%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jean-Claude%22%2C%22lastName%22%3A%22Liebart%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ula%20V.%22%2C%22lastName%22%3A%22Jurkunas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Marc%22%2C%22lastName%22%3A%22Nadal%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Philippe%22%2C%22lastName%22%3A%22Bouloc%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Julien%22%2C%22lastName%22%3A%22Dairou%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Aazdine%22%2C%22lastName%22%3A%22Lamouri%22%7D%5D%2C%22abstractNote%22%3A%22DNA%20damage%20induced%20by%20reactive%20carbonyls%20%28mainly%20methylglyoxal%20and%20glyoxal%29%2C%20called%20DNA%20glycation%2C%20is%20quantitatively%20as%20important%20as%20oxidative%20damage.%20DNA%20glycation%20is%20associated%20with%20increased%20mutation%20frequency%2C%20DNA%20strand%20breaks%2C%20and%20cytotoxicity.%20However%2C%20in%20contrast%20to%20guanine%20oxidation%20repair%2C%20how%20glycated%20DNA%20is%20repaired%20remains%20undetermined.%20Here%2C%20we%20found%20that%20the%20parkinsonism-associated%20protein%20DJ-1%20and%20its%20bacterial%20homologs%20Hsp31%2C%20YhbO%2C%20and%20YajL%20could%20repair%20methylglyoxal-%20and%20glyoxal-glycated%20nucleotides%20and%20nucleic%20acids.%20DJ-1-depleted%20cells%20displayed%20increased%20levels%20of%20glycated%20DNA%2C%20DNA%20strand%20breaks%2C%20and%20phosphorylated%20p53.%20Deglycase-deficient%20bacterial%20mutants%20displayed%20increased%20levels%20of%20glycated%20DNA%20and%20RNA%20and%20exhibited%20strong%20mutator%20phenotypes.%20Thus%2C%20DJ-1%20and%20its%20prokaryotic%20homologs%20constitute%20a%20major%20nucleotide%20repair%20system%20that%20we%20name%20guanine%20glycation%20repair.%22%2C%22date%22%3A%222017-07-14%22%2C%22section%22%3A%22%22%2C%22partNumber%22%3A%22%22%2C%22partTitle%22%3A%22%22%2C%22DOI%22%3A%2210.1126%5C%2Fscience.aag1095%22%2C%22citationKey%22%3A%22%22%2C%22url%22%3A%22%22%2C%22PMID%22%3A%22%22%2C%22PMCID%22%3A%22%22%2C%22ISSN%22%3A%221095-9203%22%2C%22language%22%3A%22eng%22%2C%22collections%22%3A%5B%22GWSKYQWE%22%5D%2C%22dateModified%22%3A%222022-09-05T13%3A28%3A17Z%22%7D%7D%5D%7D
Joly, N., & Pintard, L. (2025). Intramolecular regulation of the MT-severing enzyme Katanin prevents futile ATP hydrolysis. Journal of Cell Biology, 225(2), e202506192. https://doi.org/10.1083/jcb.202506192
Roumbo, L., Ossareh-Nazari, B., Vigneron, S., Stefani, I., Van Hove, L., Legros, V., Chevreux, G., Lacroix, B., Castro, A., Joly, N., Lorca, T., & Pintard, L. (2025). The MAST kinase KIN-4 carries out mitotic entry functions of Greatwall in C. elegans. The EMBO Journal, 1–32. https://doi.org/10.1038/s44318-025-00364-w
El Mossadeq, L., Bellutti, L., Le Borgne, R., Canman, J. C., Pintard, L., Verbavatz, J.-M., Askjaer, P., & Dumont, J. (2024). An interkinetic envelope surrounds chromosomes between meiosis I and II in C. elegans oocytes. Journal of Cell Biology, 224(3), e202403125. https://doi.org/10.1083/jcb.202403125
Strzelecki, P., Joly, N., Hébraud, P., Hoffmann, E., Cech, G. M., Kloska, A., Busi, F., & Grange, W. (2024). Enhanced Golden Gate Assembly: evaluating overhang strength for improved ligation efficiency. Nucleic Acids Research, gkae809. https://doi.org/10.1093/nar/gkae809
Beaumale, E., Van Hove, L., Pintard, L., & Joly, N. (2024). Microtubule-binding domains in Katanin p80 subunit are essential for severing activity in C. elegans. Journal of Cell Biology, 223(4), e202308023. https://doi.org/10.1083/jcb.202308023
Nkombo Nkoula, S., Velez-Aguilera, G., Ossareh-Nazari, B., Van Hove, L., Ayuso, C., Legros, V., Chevreux, G., Thomas, L., Seydoux, G., Askjaer, P., & Pintard, L. (2023). Mechanisms of nuclear pore complex disassembly by the mitotic Polo-like kinase 1 (PLK-1) in C. elegans embryos. Science Advances, 9(29), eadf7826. https://doi.org/10.1126/sciadv.adf7826
Kouranti, I., Abdel Khalek, W., Mazurkiewicz, S., Loisel-Ferreira, I., Gautreau, A. M., Pintard, L., Jeunemaitre, X., & Clauser, E. (2022). Cullin 3 Exon 9 Deletion in Familial Hyperkalemic Hypertension Impairs Cullin3-Ring-E3 Ligase (CRL3) Dynamic Regulation and Cycling. International Journal of Molecular Sciences, 23(9), 5151. https://doi.org/10.3390/ijms23095151
Velez-Aguilera, G., Ossareh-Nazari, B., Van Hove, L., Joly, N., & Pintard, L. (2022). Cortical microtubule pulling forces contribute to the union of the parental genomes in the Caenorhabditis elegans zygote. eLife, 11, e75382. https://doi.org/10.7554/eLife.75382
Knox, J., Joly, N., Linossi, E. M., Carmona-Negrón, J. A., Jura, N., Pintard, L., Zuercher, W., & Roy, P. J. (2021). A survey of the kinome pharmacopeia reveals multiple scaffolds and targets for the development of novel anthelmintics. Scientific Reports, 11(1), 9161. https://doi.org/10.1038/s41598-021-88150-6
Tavernier, N., Thomas, Y., Vigneron, S., Maisonneuve, P., Orlicky, S., Mader, P., Regmi, S. G., Van Hove, L., Levinson, N. M., Gasmi-Seabrook, G., Joly, N., Poteau, M., Velez-Aguilera, G., Gavet, O., Castro, A., Dasso, M., Lorca, T., Sicheri, F., & Pintard, L. (2021). Bora phosphorylation substitutes in trans for T-loop phosphorylation in Aurora A to promote mitotic entry. Nature Communications, 12(1), 1899. https://doi.org/10.1038/s41467-021-21922-w
Velez-Aguilera, G., Nkombo Nkoula, S., Ossareh-Nazari, B., Link, J., Paouneskou, D., Van Hove, L., Joly, N., Tavernier, N., Verbavatz, J.-M., Jantsch, V., & Pintard, L. (2020). PLK-1 promotes the merger of the parental genome into a single nucleus by triggering lamina disassembly. eLife, 9, e59510. https://doi.org/10.7554/eLife.59510
Joly, N., Beaumale, E., Van Hove, L., Martino, L., & Pintard, L. (2020). Phosphorylation of the microtubule-severing AAA+ enzyme Katanin regulates C. elegans embryo development. The Journal of Cell Biology, 219(6), e201912037. https://doi.org/10.1083/jcb.201912037
Gutnik, S., Thomas, Y., Guo, Y., Stoecklin, J., Neagu, A., Pintard, L., Merlet, J., & Ciosk, R. (2018). PRP-19, a conserved pre-mRNA processing factor and E3 ubiquitin ligase, inhibits the nuclear accumulation of GLP-1/Notch intracellular domain. Biology Open, 7(7), bio034066. https://doi.org/10.1242/bio.034066
Vigneron, S., Sundermann, L., Labbé, J.-C., Pintard, L., Radulescu, O., Castro, A., & Lorca, T. (2018). Cyclin A-cdk1-Dependent Phosphorylation of Bora Is the Triggering Factor Promoting Mitotic Entry. Developmental Cell, 45(5), 637-650.e7. https://doi.org/10.1016/j.devcel.2018.05.005
Martino, L., Morchoisne-Bolhy, S., Cheerambathur, D. K., Van Hove, L., Dumont, J., Joly, N., Desai, A., Doye, V., & Pintard, L. (2017). Channel Nucleoporins Recruit PLK-1 to Nuclear Pore Complexes to Direct Nuclear Envelope Breakdown in C. elegans. Developmental Cell, 43(2), 157-171.e7. https://doi.org/10.1016/j.devcel.2017.09.019
Dickinson, D. J., Schwager, F., Pintard, L., Gotta, M., & Goldstein, B. (2017). A Single-Cell Biochemistry Approach Reveals PAR Complex Dynamics during Cell Polarization. Developmental Cell, 42(4), 416-434.e11. https://doi.org/10.1016/j.devcel.2017.07.024
Richarme, G., Liu, C., Mihoub, M., Abdallah, J., Leger, T., Joly, N., Liebart, J.-C., Jurkunas, U. V., Nadal, M., Bouloc, P., Dairou, J., & Lamouri, A. (2017). Guanine glycation repair by DJ-1/Park7 and its bacterial homologs. Science (New York, N.Y.), 357(6347), 208–211. https://doi.org/10.1126/science.aag1095
Revues
Chapitre de livre
- Frank SICHERI (University of Toronto, Canada)
- Thierry LORCA (CRBM, Montpellier, France)
- Anna CASTRO (CRBM, Montpellier, France)
- Olivier GAVET (Institut Gustave Roussy, Paris Villejuif)
- Mary DASSO (NIH, Bethesda USA)
- Peter ASKJAER (CABD, Sevilla, Spain)
- Verena JANTSCH (Max Perutz Labs Vienna, Austria)
- Antoine JEGOU (Institut Jacques Monod, Paris, France)
- Guillaume ROMET-LEMONNE (Institut Jacques Monod, Paris, France)
- Denis CHRETIEN (IGDR, Rennes, France)
- Julien DUMONT (Institut Jacques Monod, Paris, France)
- Valérie DOYE (Institut Jacques Monod, Paris, France)
- Bruce BOWERMAN (IMB, University of Oregon, USA)
- Arshad DESAI (USCD, San Diego, USA)
- Monica GOTTA (University of Geneva, Switzerland)
- ANR AMBRE
- ANR REPLIGREAT
- ARC
- Idex « AAP Dynamique » Université de Paris
- Equipe Labellisée Ligue contre le Cancer
- Labex « WHO AM I »
26/11/2025 : Actualité scientifiqie publiée par CNRS Biologie : Un mécanisme d’auto-régulation empêche la Katanine de gaspiller l’énergie cellulaire
25/10/2021 : Nicolas JOLY (Institut Jacques Monod), lauréat du prix Maurice Nicloux 2021 !
Master and PhD students
Our laboratory offers a wide range of interesting and challenging research projects for motivated master or PhD candidates aimed at understanding the mechanisms of cell cycle control during animal development.
We employ a unique combination of genetics, biochemistry, cell biology, live imaging, quantitative proteomics, and functional genomics approaches.
PostDoc candidates
We are looking for highly motivated and team-oriented scientists with a strong background in biochemistry, genetics and cell biology.
Candidates are welcome to apply with CV, publication list, motivation letter, and names of 2 referees to:
Lionel PINTARD – Université Paris cité, CNRS, Institut Jacques Monod, 15 rue Hélène Brion – 75013 PARIS cedex 13 – France


