Chromosomal Domains and DNA Replication
MARIE-NOËLLE PRIOLEAU
Replication is a highly regulated process, which is initiated once and only once per cell cycle in all living organisms. We are interested in identifying the molecular mechanisms that implement this program and understanding how transcription and replication may process harmoniously on the same molecule. We use a large spectrum of experimental approaches (cytological, genetic, genomic, bioinformatic) and large diversity of model organisms – bacteria (Escherichia coli, Vibrio cholerae) and vertebrates (chicken and human) – to tackle these questions.
The team has research direction: replication initiation in vertebrates (Marie-Noëlle Prioleau) and replication initiation in bacteria (Jean-Luc Ferat).
Key words : DNA replication, replication initiation, G-quadruplex, Chromatin, Commun Fragile Sites, Cortical Organoïd, bacteria, replicative co-helicase
+33 (0)1 57 27 81 02 Contact @MariePrioleau
Replication initiation in vertebrates (Marie-Noëlle Prioleau)
In eukaryotes, the spatiotemporal program controls the positioning and the activation time of the origins of replication during the S phase of the cell cycle. We are. More recently, we are also trying to understand. Indeed, it is now clearly established that conflicts between these two machineries can be a source of genomic instability.
In order to get a general view of human genome replication, we are developing high-throughput analyses to map the origins of replication as well as their activation time (Figure 1).
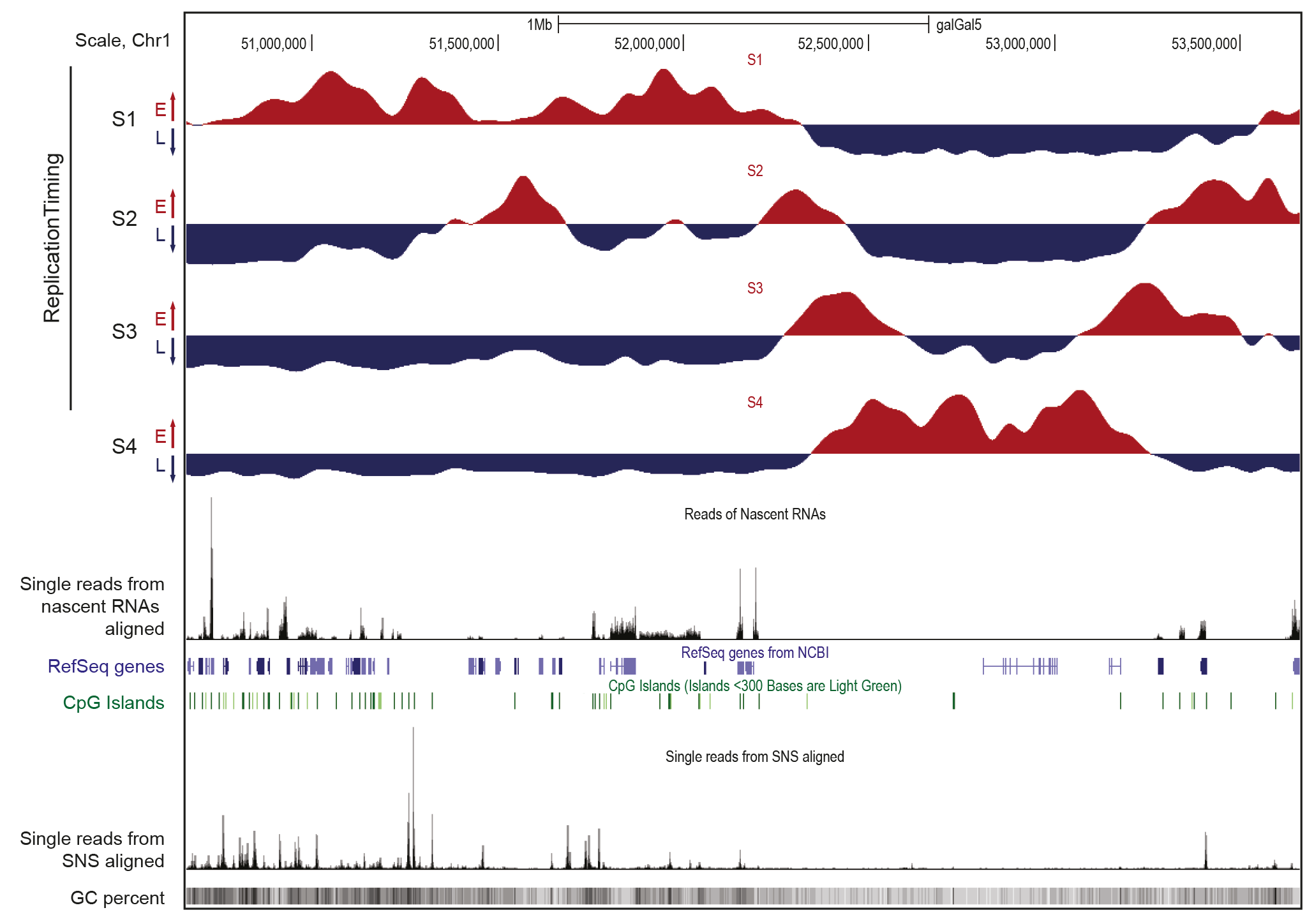 Figure 1. Representative features of early and late replication domains
Figure 1. Representative features of early and late replication domains
UCSC genome browser visualization of a 3Mb genomic region of the avian chromosome 1. Data were obtained in the DT40 avian cell line. Tracks of nascent strands (NS) enrichments in the four S-phase fractions, from early to late, are shown separately (S1 to S4). NS-enriched and depleted regions for each fraction are shown in red and blue, respectively. This data provides information on the temporal program of DNA replication. Single reads from nascent RNA-seq data and aligned SNS (spatial program of DNA replication) are reported and between tracks of annotated genes (RefSeq genes) and CpG Islands. The bottom track shows GC content (GC percent).
Collaboration with statisticians and bioinformaticians has allowed us to link these maps to genomic data on chromatin structure and gene expression. We also use an avian cell model (the DT40 cell line) which has the unique property of performing homologous recombination very efficiently. This powerful genetic model allows us to test very efficiently different hypotheses extracted from genomic analyses. We were able to show that G-quadruplexes play a key role in the definition of replication origins. We have just identified a complex motif that is found in half of the strong origins in humans, mice and chickens. Future projects will focus on understanding how this motif ensures the efficient recruitment of the replication machinery.
In a second line of research, we are analyzing how the origin signal carried by this complex motif interacts with the transcription process that takes place on the same substrate, DNA. These analyses will allow us to test a well-supported hypothesis that transcription is able to shift the starting points of replication and thus generally promote replication initiation in intergenic regions. Common Fragile Sites (CFSs) are recurrent sites of chromosomal rearrangements in cancers and some neurological diseases. They are found within large (> 300 kb) genes transcribed and replicated at the end of S phase. One hypothesis regarding their formation is that the lack of replication initiation events along these genes results in incomplete replication of these regions before mitosis. Transcription would remove the pre-replication complexes located within the bodies of these genes, thus inducing a depletion of replication origins. We seek to test the ability of RNA polymerase II to displace pre-replication complexes by inserting a strong or inducible promoter upstream of a highly efficient minimal model origin lacking transcriptional activity. This project will explore an important hypothesis on the formation of CFS. In the longer term, we will test the hypothesis that recurrent break sites observed during the proliferation of neuronal precursors have the characteristic properties of CFS. For this purpose, we will use human brain organoids as a model system.
The understanding of the duplication mode of eukaryotic genomes is essential. Indeed, replication not only ensures the maintenance of genome integrity, but also coordinates the establishment of expression programs during development.
Replication initiation in bacteria (Jean-Luc Ferat)
In bacteria, replication is initiated at a single origin by the initiator protein, DnaA. The ordered binding of DnaA at the origin leads to the local unwinding of DNA and the direct recruitment of replicative helicases on single-strand DNA. This step is highly regulated and assisted by a co-helicase, which synchronizes the loading of the replicative helicases, ensuring Bidirectional Replication Initiation (BRI). We are interested in understanding 1) how does the co-helicase DciA load and activate synchronously the replicative helicases during replication initiation and 2) how does a replication initiated in absence of DciA result in a pathological degradation of the whole chromosome. We developed a large spectrum of experimental approaches (cytological, genetic, genomic, bioinformatic) to address these two questions in the model organisms Vibrio cholerae and Escherichia coli.
In bacteria, replication is initiated at a single origin by the initiator protein, DnaA. The ordered binding of DnaA at the origin creates a local unwinding of the double helix that enables the recruitment of the replicative helicase, DnaB. Replisomes are then assembled on this nucleoprotein platform to trigger Bidirectional Replication Initiation. Contrary to eukaryotes, bacteria load directly their helicase on single strand DNA during replication initiation. This process is highly regulated and assisted by co-helicases in the three domains of life. Bacterial co-helicases are diverse. The first family that was studied, DnaC/I (DnaC in Escherichia coli and DnaI in Bacillus subtilis), is present in only a few orders of bacteria and results from the domestication of a phage gene (Brézellec et al., 2016). The main family of co-helicase is DciA. It was discovered only a few years ago although it is present in the vas majority of bacterial orders (Figure 1). We established that DciA, is the ancestral bacterial co-helicase (Brézellec et al., 2016, 2017).
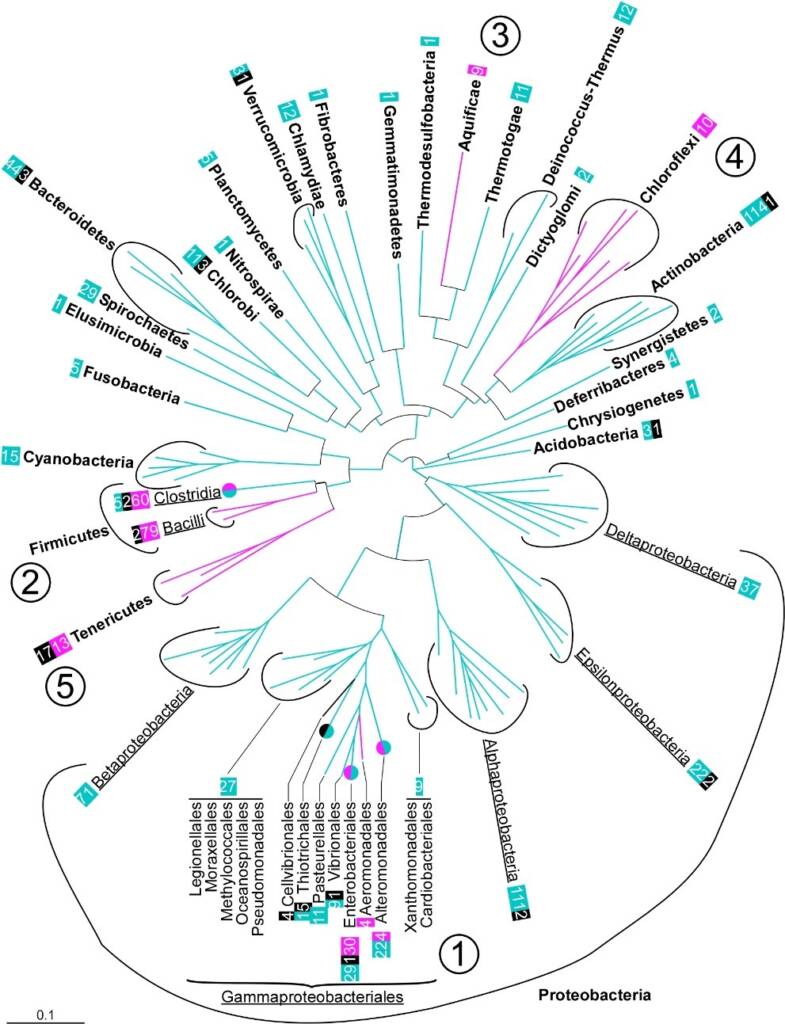
Fig. 1 Distribution of domesticated dnaC and dciA in the bacterial domain. Distribution of the bacterial phyla investigated on a 16S rRNA-based circular phylogenetic tree (Methods). When the presence of dnaC/I or dciA within a phylum (bold) was not homogeneous, classes (underlined) or orders (normal) are specified. Numbers boxed refer to the number of species’ genomes (and not strains) containing domesticated dnaC/I (fuchsia), dciA (turquoise) or neither (black). fuchsia / turquoise circles: orders in which dciA and dnaC/I genes were identified. The order Thiotrichales is identified with a green/black circle because most species lack dciA and dnaC/I. ① Gammaproteobacteria, ② Firmicutes, ③ Aquificae, ④ Chloroflexi and ⑤ Tenericutes.
We conducted a biochemical investigation of DciA together with its cognate replicative helicase to compare its activity to that of DnaC of E. coli. This study revealed surprisingly that contrary to DnaC/I the loading of the replicative helicase is not dependent on DciA. In other words, the replicative helicase of dciA-sepcies can self-load on DNA. At most, DciA improves the loading efficiency (Figure 2) (Marsin et al., 2021).
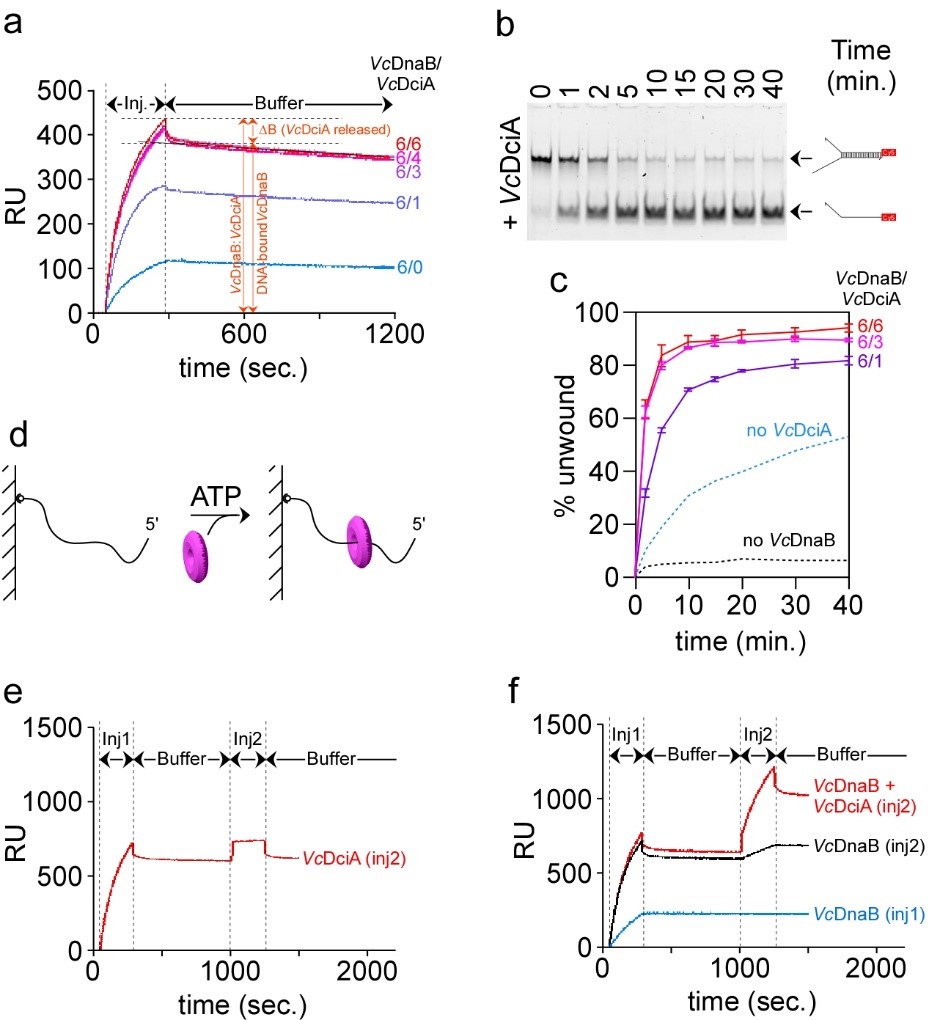
Fig. 2 VcDciA stimulates the loading of VcDnaB on DNA. a, SPR analysis of the loading of VcDnaB on DNA in presence of VcDciA. Experiments were conducted as described in Figure 1B in presence of VcDnaB at 625nM. Different VcDnaB/VcDciA ratios (from 6/0 to 6/6 in subunits) were analyzed. ΔB represents the mass of VcDciA proteins that dissociated immediately after the loading of the VcDciA•VcDnaB complex on DNA. b, DNA unwinding kinetics in presence of VcDciA. The experiments were carried out as described in Figure 1D, except that VcDciA (variable concentrations) was added to VcDnaB (625 nM). The types of migration products are indicated on the right. c, DNA unwinding quantification in presence of VcDciA. The procedure is the same as that described in Figure 1E. The error bars correspond to the mean +/- SEM of three independent experiments. d, Helicase loading in absence of Mg2+. The experiment is carried out in absence of Mg to prevent the activation of the helicase and its translocation on DNA in the 5’ → 3’ direction until its release. ATP was added for it is required for the hexamerization of the helicase.
Flow cytometry analysis of [dciA–] cells confirmed that the replicative helicase of V. cholerae can load itself in vivo without DciA. A genomic approach intended to assessing the replication status of [dciA–] cells revealed that DciA is essential to ensure Bidirectional Replication Initiation (BRI), likely by synchronizing the loading of the replicative helicase during replication initiation. We showed that in absence of DciA, replication is initiated unidirectionally and that this Unidirectional Replication Initiation (URI) is infallibly followed by a catastrophic sequence of events ending in chromosome degradation (Besombes et al. submitted) (Figure 3).
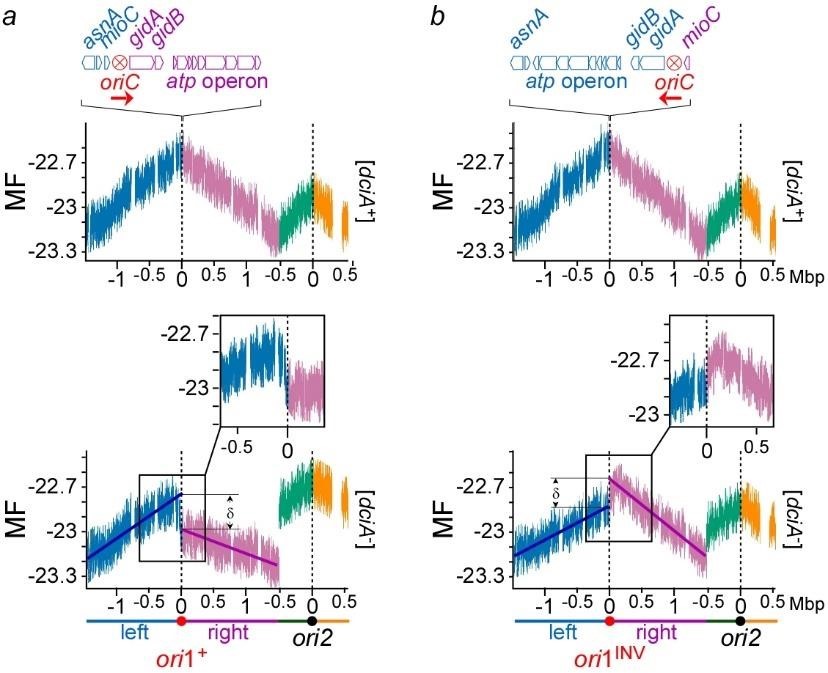
Fig.3 Replication is initiated unidirectionally at ori1 on Chr1 in [dciA–] cells. a) MF profiles of ori1+ dciA+ (top )and ori1+ [dciA–] populations (bottom) maintained in continuous growth in minimal medium at 30°C for 12h before DNA extraction and sequence. The reads were aligned against V. cholerae reference genome (using Burrows-Wheeler Aligner) to assign a number of reads per genome position (marker frequency). The value for each position of the genome corresponds to the marker frequency average of 1000 consecutive positions (1kb) surrounding it. log2 of these values are used to correct for the exponential growth of the cells (MF). b) same as in a), except that the origin of replication was inverted (oriINV). The left and right replichore of Chr1 and Chr2 are colored in blue, purple, green and orange, respectively.
Our research program is aimed now at understanding how unprogrammed URI leads to chromosome degradation and how DciA loads and activates synchronously the replicative helicases during replication initiation. We address these questions 1) by inventorying the diverse routes followed by [dciA–] cells and dissecting them frame by frame through a large diversity of approaches (fluorescent microscopy, digital PCR, PFGE, NGS, etc.), 2) by deciphering the functional network of DciA (two hybrid, Tn-seq) and 3) by creating a strain of V. cholerae in which dciA is replaced by dnaC to trace the modifications and their succession during adaptation to a new co-helicase and eventually to understand the mechanism underlying BRI.
We use two bacterial model organisms (Vibrio cholerae and Escherichia coli) to address these two questions, and developed a large spectrum of experimental approaches (cytological, genetic, genomic, bioinformatic).
References
Besombes A, Adam Y, Possoz C, Junier Y, Barre FX, Ferat JL. (submitted) Bidirectional replication initiation prevents chromosome degradation
Marsin S, Adam Y, Cargemel C, Andreani J, Baconnais S, Legrand P, Li de la Sierra-Gallay I, Humbert A, Aumont-Nicaise M, Velours C, Ochsenbein F, Durand D, Le Cam E, Walbott H, Possoz C, Quevillon-Cheruel S, Ferat JL. (2021) Study of the DnaB:DciA interplay reveals insights into the primary mode of loading of the bacterial replicative helicase. Nucleic Acids Res. 49:6569-6586
Brézellec P, Petit MA, Pasek S, Vallet-Gely I, Possoz C, Ferat JL. (2017) Domestication of Lambda Phage Genes into a Putative Third Type of Replicative Helicase Matchmaker. Genome Biol Evol. 9:1561-1566
Brézellec P, Vallet-Gely I, Possoz C, Quevillon-Cheruel S, Ferat JL. (2016) DciA is an ancestral replicative helicase operator essential for bacterial replication initiation. Nat Commun. 7:13271
Members
 Theo BARET, Ingénieur de recherche en bioinformatique génomique, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B
Theo BARET, Ingénieur de recherche en bioinformatique génomique, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B Amélie BESOMBES, Doctorante, PRIOLEAU LABroom 520B
Amélie BESOMBES, Doctorante, PRIOLEAU LABroom 520B Thibault CORNELOUP, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B
Thibault CORNELOUP, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B Caroline DONCARLI, Ingénieure en biologie, PRIOLEAU LAB+33 (0)1 57 27 81 24, room 520B
Caroline DONCARLI, Ingénieure en biologie, PRIOLEAU LAB+33 (0)1 57 27 81 24, room 520B Jean-Luc FERAT, Enseignant-chercheur, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B
Jean-Luc FERAT, Enseignant-chercheur, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B Juliette MANDELBROJT, Doctorante, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B
Juliette MANDELBROJT, Doctorante, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B Kathrin MARHEINEKE, Chercheur, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B
Kathrin MARHEINEKE, Chercheur, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B Aurélie MASSON, Doctorante, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B
Aurélie MASSON, Doctorante, PRIOLEAU LAB+33 (0)1 57 27 81 02, room 520B
To contact a member of the team by e-mail: name.surname@ijm.fr
1) Clustering of strong replicators associated with active promoters is sufficient to establish an early-replicating domain. Brossas C, Valton AL, Venev SV, Chilaka S, Counillon A, Laurent M, Goncalves C, Duriez B, Picard F, Dekker J, Prioleau MN. EMBO J. 2020 Nov 2;39(21):e99520. doi: 10.15252/embj.201899520. Epub 2020 Sep 16. PMID: 32935369
2) Replication dynamics of individual loci in single living cells reveal changes in the degree of replication stochasticity through S phase. Duriez B, Chilaka S, Bercher JF, Hercul E, Prioleau MN. Nucleic Acids Res. 2019 Jun 4;47(10):5155-5169. doi: 10.1093/nar/gkz220. PMID: 30926993
3) Evolution of replication origins in vertebrate genomes: rapid turnover despite selective constraints. Massip F, Laurent M, Brossas C, Fernández-Justel JM, Gómez M, Prioleau MN, Duret L, Picard F. Nucleic Acids Res. 2019 Jun 4;47(10):5114-5125. doi: 10.1093/nar/gkz182. PMID: 30916335
4) Transcription-dependent regulation of replication dynamics modulates genome stability. Blin M, Le Tallec B, Nähse V, Schmidt M, Brossas C, Millot GA, Prioleau MN, Debatisse M. Nat Struct Mol Biol. 2019 Jan;26(1):58-66. doi: 10.1038/s41594-018-0170-1. Epub 2018 Dec 31. PMID: 30598553
5) The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. Picard F, Cadoret JC, Audit B, Arneodo A, Alberti A, Battail C, Duret L, Prioleau MN. PLoS Genet. 2014 May 1;10(5):e1004282. doi: 10.1371/journal.pgen.1004282. eCollection 2014 May. PMID: 24785686
6) G4 motifs affect origin positioning and efficiency in two vertebrate replicators. Valton AL, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintomé C, Riou JF, Prioleau MN. EMBO J. 2014 Apr 1;33(7):732-46. doi: 10.1002/embj.201387506. Epub 2014 Feb 12. PMID: 24521668
7) USF binding sequences from the HS4 insulator element impose early replication timing on a vertebrate replicator. Hassan-Zadeh V, Chilaka S, Cadoret JC, Ma MK, Boggetto N, West AG, Prioleau MN. PLoS Biol. 2012;10(3):e1001277. doi: 10.1371/journal.pbio.1001277. Epub 2012 Mar 6. PMID: 22412349
8) Genome-wide studies highlight indirect links between human replication origins and gene regulation. Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN. Proc Natl Acad Sci U S A. 2008 Oct 14;105(41):15837-42. doi: 10.1073/pnas.0805208105. Epub 2008 Oct 6. PMID: 18838675
9) Broadening of DNA replication origin usage during metazoan cell differentiation. Dazy S, Gandrillon O, Hyrien O, Prioleau MN. EMBO Rep. 2006 Aug;7(8):806-11. doi: 10.1038/sj.embor.7400736. Epub 2006 Jun 16. PMID: 16799461
10) Replication of the chicken beta-globin locus: early-firing origins at the 5′ HS4 insulator and the rho- and betaA-globin genes show opposite epigenetic modifications. Prioleau MN, Gendron MC, Hyrien O. Mol Cell Biol. 2003 May;23(10):3536-49. doi: 10.1128/MCB.23.10.3536-3549.2003. PMID: 12724412
Publications
Reviews
- « Rôle de l’insulateur 5’HS4 du poulet dans la régulation de la réplication » soutenue en Juin 2009 par Vahideh Hassan-Zadeh.
- « Mécanismes moléculaires de l’initiation de la réplication » soutenue en Décembre 2010 par Françoise Meisch.
- « Identification de séquences cis-nucléiques nécessaires à l’initiation de la réplication chez les vertébrés » soutenue en Juin 2014 par Anne-Laure Valton.
- « Construction d’un domaine synthétique de réplication précoce et impact sur la structure chromatinienne et la permissivité transcriptionnelle » soutenue en Septembre 2015 par Caroline Tonnerre-Doncarli Brossas.
- « Mécanismes moléculaires impliqués dans la régulation du moment de déclenchement des origines de réplication » soutenue en Novembre 2016 par Antonin Counillon.
- « Rôle des G-quadruplexes dans la spécification des origines de réplication chez les vertébrés » soutenue en Septembre 2016 par Marc Laurent.
- « Etude moléculaire des éléments cis-régulateurs et de l’organisation de la chromatine des origines de réplication » soutenue en Septembre 2020 par Jérémy Poulet-Benedetti.
Nationals:
- Imagerie cellulaire et cytométrie en flux : Plateforme Imago-Seine, Institut Jacques Monod
- Organoïdes corticaux : Plateforme enSCORE, Labex Who am I ?
- Bio-informatique et bio-statistique : LBBE, UCB Lyon 1 (Laurent Duret) et ENS de Lyon (Franck Picard)
- G-quadruplex : Inserm U565, CNRS UMR 7196, MNHN, Paris, France (Patrizia Alberti, Carole Saintomé et Jean-François Riou)
- Sites Fragiles Communs : : Institut Gustave Roussy, Villejuif (Michelle Debatisse et Stéphane Koundrioukoff)
Internationals:
- Organisation tri-dimensionnelle du noyau : University of Massachusetts Medical School, Worcester, Etats-Unis (Anne-Laure Valton et Job Dekker)
- Rôle des G4s dans le positionnement des nucléosomes : Fox Chase Cancer center, Philadelphie, Etats-Unis et Moscow State University, Moscow, Russie (Vasily Studitsky).
Equipe-partenaire du Labex “Who am I?”


